Identification of calmodulin (ScCaM) gene and its correlation with shell calcium carbonate deposition of Sinonovacula constricta
-
摘要: 碳酸钙(CaCO3)作为贝壳的主要成分,与有机质框架相互作用形成贝壳,为贝类提供保护作用。Ca2+是CaCO3的重要组成部分,其在贝类体内的获取、转运、沉淀过程均会显著影响CaCO3沉积。然而,目前关于贝壳中碳酸钙沉积过程以及相关基因的作用机制仍不明晰。钙调蛋白(Calmodulin, CaM)是一种广泛存在于真核细胞中并与Ca2+特异性结合的蛋白,主要参与细胞信号转导、靶酶活性调控和Ca2+稳态调节等多种生理过程。为研究CaM基因与贝壳碳酸钙沉积的关系,本研究对缢蛏CaM基因(ScCaM)进行分子鉴定和表达特征分析,并研究了ScCaM重组蛋白的Ca2+结合活性及其在CaCO3沉积中的作用。结果显示,ScCaM基因共编码149个氨基酸,含有4个连续EF-hand结构域;ScCaM在各组织中均能表达,其中在鳃和外套膜组织中表达水平显著高于足、水管、闭壳肌和肝胰腺等组织(P < 0.05);缢蛏壳碳酸钙含量与其壳重呈正比,并且贝壳重量较大的个体,其ScCaM基因表达水平较高。ScCaM重组蛋白具有钙离子结合活性,可加快碳酸钙沉淀速率,并且促进效应呈现出明显的蛋白浓度依赖性。结果表明,ScCaM基因/蛋白表达与贝壳碳酸钙含量密切相关,即ScCaM基因/蛋白表达量升高,可增强Ca2+运输效率,促进壳碳酸钙沉积,从而提高贝壳重量。本研究初步探讨了ScCaM基因在贝壳碳酸钙沉积中的作用,为解析缢蛏贝壳形成的分子机制提供了理论基础。Abstract: Calcium carbonate (CaCO3), as a major component of shells, interacts with the organic matter framework to form shells and provide protection for mollusks. Ca2+ is an important component of CaCO3, and its acquisition, transport, and precipitation processes can significantly affect calcium carbonate deposition in mollusks. However, there is still a lack of clarity regarding the process of calcium carbonate deposition and mechanism of related genes in forming shells. Calmodulin (CaM) is a protein widely found in eukaryotic cells and specifically binds to Ca2+, which is mainly involved in a variety of physiological processes such as cellular signal transduction, regulation of target enzyme activities and regulation of Ca2+ homeostasis. In order to investigate the relationship between CaM gene and calcium carbonate deposition in shells, we performed molecular identification and expression characterization of CaM gene in Sinonovacula constricta (ScCaM), and investigated the Ca2+-binding activity of the recombinant protein ScCaM and its role in calcium carbonate deposition. The results showed that the ScCaM gene encoded a total of 149 amino acids and contained four consecutive EF-hand structural domains. ScCaM was expressed in all tissues, with the expression level in gill and mantle tissues being significantly higher than in foot, siphon, adductor muscle and hepatopancreas tissues (P < 0.05). Furthermore, the content of calcium carbonate in shells was positively proportional to their shell weight. Meanwhile, individuals with larger shell weights had higher expression levels of ScCaM. ScCaM recombinant protein had calcium ion binding activity, which can accelerate the rate of calcium carbonate deposition, and the promotion effect showed an obvious protein concentration dependence. The results showed that ScCaM gene/protein expression was closely related to shell calcium carbonate content: elevated ScCaM gene/protein expression could enhance Ca2+ transport efficiency, promote shell calcium carbonate deposition, and thus increase shell weight. This study preliminarily investigated the role of ScCaM gene in shell calcium carbonate deposition, and provided a theoretical basis for analyzing the molecular mechanism of shell formation in S. constricta.
-
Key words:
- Sinonovacula constricta /
- calmodulin (CaM) /
- shell weight /
- calcium carbonate /
- expression
-
图 1 CaM蛋白的氨基酸多序列比对(A)、同源性分析(B)、邻接法构建系统进化树(C)和蛋白三级结构(D)
注:红色三角标记为缢蛏CaM特有氨基酸残基,红色方框标注为氨基酸多态性。蛋白三级结构各结构域以相同颜色色块标注于序列比对图中。
Fig. 1 Multiple amino acid sequence alignment of CaM proteins (A), homology analysis among CaM proteins (B), phylogenetic tree constructed by neighbour-joining method (C), and tertiary structure of CaM protein (D)
Note: Red triangle indicates specific amino acid residues in the CaM protein of S.constricta, and red box indicates amino acid polymorphism sites. The structural domains of the protein tertiary structure are annotated with the same color blocks in Fig. 1(A).
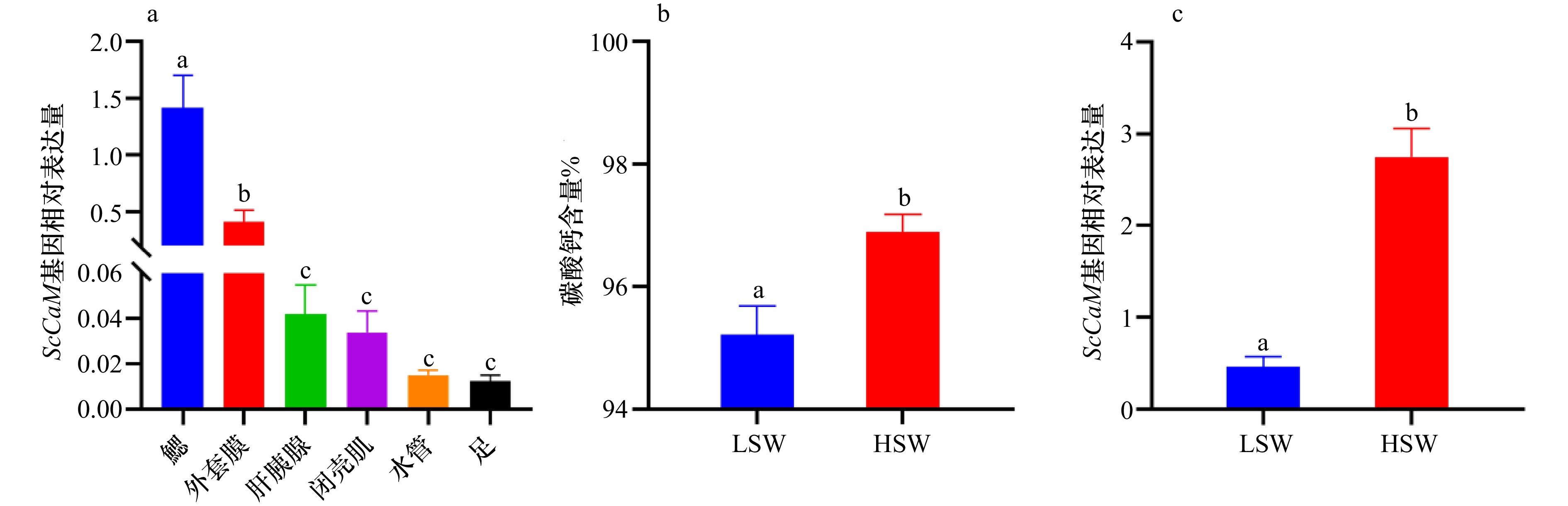
图 2 ScCaM基因在不同组织中的表达(A,n=6)、不同壳重组碳酸钙含量(B,n=30)和ScCaM基因表达(C,n=6)分析
注:不同小写字母表示差异显著 (P < 0.05);LSW:低贝壳重量组;HSW:高贝壳重量组
Fig. 2 The relative expression level of ScCaM in various tissues (A, n=6), the calcium carbonate content (B, n=30) and ScCaM gene expression level (C, n=6) analysis in different shell weight groups.
Note: Different lowercase letters indicate significant differences (P < 0.05); LSW: low shell weight group; HSW: high shell weight group.
图 3 ScCaM基因的原核表达和蛋白纯化分析
注:A. 重组钙调蛋白原核表达: a. pET-32a空载,b. pET-32a-CaM上清,c. pET-32a-CaM沉淀,d. pET-32a-CaM全菌,M. Marker;B. 缢蛏钙调蛋白原核表达纯化: 1. pET-32a空载,2. pET-32a空载+IPTG,3. pET-32a-CaM,4. pET-32a-CaM+IPTG,5. pET-32a-CaM纯化;红框表示重组蛋白rScCaM
Fig. 3 Analysis of prokaryotic expression and protein purification of ScCaM gene
Note: A. Recombinant calmodulin prokaryotic expression results: a. pET-32a unloaded; b. pET-32a-CaM supernatant; c. pET-32a-CaM precipitation, d. pET-32a-CaM whole bacteria; M. protein marker, B. Results of purification of prokaryotic expression of S. constricta calmodulin: 1. pET-32a unloaded, 2. pET-32a unloaded + IPTG, 3. pET-32a-CaM, 4. pET-32a-CaM + IPTG, 5. pET-32a-CaM purification. Red boxes indicates recombinant protein rScCaM.
图 4 ScCaM重组蛋白的钙离子结合活性(A)及碳酸钙沉积速率分析(B)
注:A:1. rScCaM + EDTA;2. rScCaM + EDTA + MgCl2;3. rScCaM + EDTA + CaCl2;4. 空白对照rScCaM. “*”表示与PBS对照组比较差异显著 (P < 0.05)
Fig. 4 Analysis of the calcium ion binding activity of ScCaM recombinant protein (A) and its influence on the rate of calcium carbonate deposition (B)
Note: A: 1. rScCaM + EDTA; 2. rScCaM + EDTA + MgCl2; 3. rScCaM + EDTA + CaCl2; 4. blank control rScCaM. “*” indicates a significant difference compared with the PBS control group (P < 0.05).
表 1 实验所用的引物和序列
Tab. 1 Primers and sequences used in this study
引物 序列(5’-3’) 用途 CaM-F TTCTTCTGTTAGTTGATCAGCCAT CDS全长验证 CaM-R ATGGCTGATCAACTAACAGAAGAAC cCaM-F GCCATGGCTGATATCGGATCCATGGCT
GATCAACTAACAGAAGAACA克隆 cCaM-R GTGGTGGTGGTGGTGCTCGAGCTACC
GCGATGTCATCATCTTCAqRTCaM-F CGACGGTAACGGCACGATAG 荧光定量 qRTCaM-R CTTCCCGGATCATTTCATCAACC RS9-R CGTCTCAAAAGGGCATTACC RS9-F TGAAGTCTGGCGTGTCAAGT -
[1] Currey J D. Mechanical properties of mother of pearl in tension[J]. Proceedings of the Royal Society B: Biological Sciences, 1977, 196(1125): 443−463. [2] 段婷婷, 郑威, 黄玉松, 等. 鲍鱼壳的跨尺度结构及性能表征[J]. 暨南大学学报(自然科学与医学版), 2018, 39(2): 105−111.Duan Tingting, Zheng Wei, Huang Yusong, et al. The multiscale structure and performance characteristics of abalone shell[J]. Journal of Jinan University (Natural Science & Medicine Edition), 2018, 39(2): 105−111. [3] Gilbert P U P A, Bergmann K D, Boekelheide N, et al. Biomineralization: integrating mechanism and evolutionary history[J]. Science Advances, 2022, 8(10): eabl9653. doi: 10.1126/sciadv.abl9653 [4] Suzuki M, Murayama E, Inoue H, et al. Characterization of Prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata)[J]. Biochemical Journal, 2004, 382(1): 205−213. doi: 10.1042/BJ20040319 [5] Xie Jun, Liang Jian, Sun Juan, et al. Influence of the extrapallial fluid of Pinctada fucata on the crystallization of calcium carbonate and shell biomineralization[J]. Crystal Growth & Design, 2016, 16(2): 672−680. [6] 李云娟. 池蝶蚌外套膜组织学及钙调蛋白基因的克隆和表达特性分析[D]. 南昌: 南昌大学, 2008.Li Yunjuan. Observation on the mantle, cloning and expression analysis of the calmodulin cDNA from Hyriopsis schlegeli[D]. Nanchang: Nanchang University, 2008. [7] Haiech J, Moulhaye S B M, Kilhoffer M C. The EF-Handome: combining comparative genomic study using FamDBtool, a new bioinformatics tool, and the network of expertise of the European Calcium Society[J]. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 2004, 1742(1/3): 179−183. [8] 徐友涵. 钙调蛋白的结构与功能(上)[J]. 生物化学与生物物理进展, 1985(1): 22−27.Xu Youhan. The structure and function of calmodulin (above)[J]. Progress in Biochemistry and Biophysics, 1985(1): 22−27. (查阅网上资料, 未找到对应的英文翻译, 请确认) [9] Stevens F C. Calmodulin: an introduction[J]. Canadian Journal of Biochemistry and Cell Biology, 1983, 61(8): 906−910. doi: 10.1139/o83-115 [10] Zhou Xiaoying, Cui Yazhou, Luan Jing, et al. Label-free quantification proteomics reveals novel calcium binding proteins in matrix vesicles isolated from mineralizing Saos-2 cells[J]. BioScience Trends, 2013, 7(3): 144−151. [11] Zhang Liang, Feng Xu, McDonald J M. The role of calmodulin in the regulation of osteoclastogenesis[J]. Endocrinology, 2003, 144(10): 4536−4543. doi: 10.1210/en.2003-0147 [12] Bhasker T V, Gowda N K S, Mondal S, et al. Boron supplementation influences bone mineralization by modulating expression of genes regulating calcium utilization[J]. Animal Nutrition and Feed Technology, 2017, 17(2): 201−215. doi: 10.5958/0974-181X.2017.00021.X [13] Ge Meiling, Mo Jing, Ip J C H, et al. Adaptive biomineralization in two morphotypes of Sternaspidae (Annelida) from the Northern China Seas[J]. Frontiers in Marine Science, 2022, 9: 984989. doi: 10.3389/fmars.2022.984989 [14] Shi Yaohua, Yu Chengcheng, Gu Zhifeng, et al. Characterization of the pearl oyster (Pinctada martensii) mantle transcriptome unravels biomineralization genes[J]. Marine Biotechnology, 2013, 15(2): 175−187. doi: 10.1007/s10126-012-9476-x [15] Li Shuo, Xie Liping, Zhang Cen, et al. Cloning and expression of a pivotal calcium metabolism regulator: calmodulin involved in shell formation from pearl oyster (Pinctada fucata)[J]. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 2004, 138(3): 235−243. doi: 10.1016/j.cbpc.2004.03.012 [16] Peng Kou, Liu Fanglan, Wang Junhua, et al. Calmodulin highly expressed during the formation of pearl sac in freshwater pearl mussel (Hyriopsis schlegelii)[J]. Thalassas: An International Journal of Marine Sciences, 2018, 34(1): 219−225. doi: 10.1007/s41208-017-0054-x [17] Chen Yige, Yao Yuanbin, Shen Xiaoya, et al. Transcription profiling reveals co-regulation mechanism of gene expression related to growth and mineralization induced by pearl cultivation in Hyriopsis cumingii[J]. Frontiers in Marine Science, 2024, 11: 1443863. doi: 10.3389/fmars.2024.1443863 [18] 尚朝, 施杨, 李文娟, 等. 不同Ca2+浓度养殖环境下三角帆蚌外套膜和内脏团组织钙调蛋白基因的表达[J]. 生物技术通报, 2016, 32(12): 152−159.Shang Chao, Shi Yang, Li Wenjuan, et al. Expressions of calmodulin gene in Hyriopsis cumingii mantle and visceral tissues under the culture environment with different Ca2+ concentrations[J]. Biotechnology Bulletin, 2016, 32(12): 152−159. [19] Zhan Xin, Gu Zhifeng, Yu Chengcheng, et al. Expressed sequence tags 454 sequencing and biomineralization gene expression for pearl sac of the pearl oyster, Pinctada fucata martensii[J]. Aquaculture Research, 2015, 46(3): 745−758. doi: 10.1111/are.12227 [20] Richards M, Xu Wei, Mallozzi A, et al. Production of calcium-binding proteins in Crassostrea virginica in response to increased environmental CO2 concentration[J]. Frontiers in Marine Science, 2018, 5: 203. doi: 10.3389/fmars.2018.00203 [21] Teixidó N, Caroselli E, Alliouane S, et al. Ocean acidification causes variable trait-shifts in a coral species[J]. Global Change Biology, 2020, 26(12): 6813−6830. doi: 10.1111/gcb.15372 [22] Sleight V A, Thorne M A S, Peck L S, et al. Transcriptomic response to shell damage in the Antarctic clam, Laternula elliptica: time scales and spatial localisation[J]. Marine Genomics, 2015, 20: 45−55. doi: 10.1016/j.margen.2015.01.009 [23] Nogueira D J, Mattos J J, Dybas P R, et al. Effects of phenanthrene on early development of the Pacific oyster Crassostrea gigas (Thunberg, 1789)[J]. Aquatic Toxicology, 2017, 191: 50−61. doi: 10.1016/j.aquatox.2017.07.022 [24] Faas G C, Raghavachari S, Lisman J E, et al. Calmodulin as a direct detector of Ca2+ signals[J]. Nature Neuroscience, 2011, 14(3): 301−304. doi: 10.1038/nn.2746 [25] 农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 2024中国渔业统计年鉴[M]. 北京: 中国农业出版社, 2024.Ministry of Agriculture and Rural Affairs of the People’s Republic of China, National Fisheries Technology Extension Center, China Society of Fisheries. China Fishery Statistical Yearbook[M]. Beijing: China Agriculture Press, 2024. [26] Lombardi S A, Chon G D, Lee J J W, et al. Shell hardness and compressive strength of the eastern oyster, Crassostrea virginica, and the Asian oyster, Crassostrea ariakensis[J]. The Biological Bulletin, 2013, 225(3): 175−183. doi: 10.1086/BBLv225n3p175 [27] Hamilton D J, Nudds T D, Neate J. Size-selective predation of blue mussels (Mytilus edulis) by common eiders (Somateria mollissima) under controlled field conditions[J]. The Auk, 1999, 116(2): 403−416. doi: 10.2307/4089374 [28] Parveen S, Chakraborty A, Chanda D K, et al. Microstructure analysis and chemical and mechanical characterization of the shells of three freshwater snails[J]. ACS Omega, 2020, 5(40): 25757−25771. doi: 10.1021/acsomega.0c03064 [29] 莫天宝, 徐洪强, 何京, 等. 五种双壳贝类贝壳微观结构观察与成分分析[J]. 海洋科学, 2022, 46(12): 41−49.Mo Tianbao, Xu Hongqiang, He Jing, et al. Microstructure and composition analysis of five species of economic bivalves[J]. Marine Sciences, 2022, 46(12): 41−49. [30] Waterhouse A M, Procter J B, Martin D M A, et al. Jalview Version 2-a multiple sequence alignment editor and analysis workbench[J]. Bioinformatics, 2009, 25(9): 1189−1191. doi: 10.1093/bioinformatics/btp033 [31] Clewley J P, Arnold C. MEGALIGN: the multiple alignment module of LASERGENE[M]//Swindell S R. Sequence Data Analysis Guidebook. Totowa: Springer, 1997: 119−129. [32] 徐海龙, 王芮, 许莉莉, 等. 6种海洋双壳类贝壳中碳酸钙、碳酸镁含量的测定[J]. 海洋通报, 2016, 35(4): 421−426. doi: 10.11840/j.issn.1001-6392.2016.04.009Xu Hailong, Wang Rui, Xu Lili, et al. Determination of calcium carbonate and magnesium carbonate in the shell of six marine bivalves[J]. Marine Science Bulletin, 2016, 35(4): 421−426. doi: 10.11840/j.issn.1001-6392.2016.04.009 [33] 周景道, 钱智光, 田芳, 等. 建议使用紫脲酸铵——萘酚绿B新混合指示剂[J]. 淮北煤师院学报, 1995, 16(3): 25−27.Zhou Jingdao, Qian Zhiguang, Tian Fang, et al. Naphthalene phenol green as new nixed indicator[J]. Journal of Huaibei Coal Mining Teachers College, 1995, 16(3): 25−27. [34] Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the $ 2^{-\Delta \Delta C_{\mathrm{T}}} $ method[J]. Methods, 2001, 25(4): 402−408. doi: 10.1006/meth.2001.1262[35] 辛晓雨. 海洋酸化下长牡蛎外套膜对钙质壳形成的调控及机制的初探[D]. 大连: 大连海洋大学, 2023.Xin Xiaoyu. Preliminary study on the regulation of calcareous shell formation by mantle under ocean acidification and its mechanism in oyster Crassostrea gigas[J]. Dalian: Dalian Ocean University, 2023. [36] 熊新威. 马氏珠母贝珍珠层基质蛋白的相互作用模式及其功能研究[D]. 湛江: 广东海洋大学, 2022.Xiong Xinwei. Interaction patterns of nacre matrix proteins and its functional studies in Pinctada fucata martensii[D]. Zhanjiang: Guangdong Ocean University, 2022. [37] 张明霞, 张科, 袁凤娟, 等. 背角无齿蚌钙调蛋白基因的克隆及Ca2+和Cd2+对其表达的影响[J]. 生态毒理学报, 2021, 16(3): 227−238. doi: 10.7524/AJE.1673-5897.20200423001Zhang Mingxia, Zhang Ke, Yuan Fengjuan, et al. Characterization of AwCaM1 from freshwater clam Anodonta woodiana and effect of Ca2+ and Cd2+ on its expressions[J]. Asian Journal of Ecotoxicology, 2021, 16(3): 227−238. doi: 10.7524/AJE.1673-5897.20200423001 [38] 李素萍, 喻达辉, 李应清, 等. 香港牡蛎钙调蛋白基因的克隆和组织表达分析[J]. 安徽农业科学, 2022, 50(24): 75−81. doi: 10.3969/j.issn.0517-6611.2022.24.019Li Suping, Yu Dahui, Li Yingqing, et al. Molecular cloning and expression analysis of Calmodulin gene in Crassostrea hongkongensis[J]. Journal of Anhui Agricultural Sciences, 2022, 50(24): 75−81. doi: 10.3969/j.issn.0517-6611.2022.24.019 [39] Ivanina A V, Borah B, Rimkevicius T, et al. The role of the vascular endothelial growth factor (VEGF) signaling in biomineralization of the oyster Crassostrea gigas[J]. Frontiers in Marine Science, 2018, 5: 309. doi: 10.3389/fmars.2018.00309 [40] Li Shuo, Xie Liping, Ma Zhuojun, et al. cDNA cloning and characterization of a novel calmodulin-like protein from pearl oyster Pinctada fucata[J]. The FEBS Journal, 2005, 272(19): 4899−4910. doi: 10.1111/j.1742-4658.2005.04899.x [41] Stommel E W, Stephens R E, Masure H R, et al. Specific localization of scallop gill epithelial calmodulin in cilia[J]. The Journal of Cell Biology, 1982, 92(3): 622−628. doi: 10.1083/jcb.92.3.622 [42] Brini M, Carafoli E, Calì T. The plasma membrane calcium pumps: focus on the role in (neuro)pathology[J]. Biochemical and Biophysical Research Communications, 2017, 483(4): 1116−1124. doi: 10.1016/j.bbrc.2016.07.117 [43] Zeng L G, Wang J H, Li Y J, et al. Molecular characteristics and expression of calmodulin cDNA from the freshwater pearl mussel, Hyriopsis schlegelii[J]. Genetics and Molecular Research, 2012, 11(1): 42−52. doi: 10.4238/2012.January.9.5 [44] 赵鲁苹, 徐焕志, 陈东, 等. 厚壳贻贝贝壳的微结构及光谱分析[J]. 浙江大学学报(理学版), 2015, 42(3): 339−346.Zhao Luping, Xu Huanzhi, Chen Dong, et al. Microstructure and spectral analysis of Mytilus coruscus shell[J]. Journal of Zhejiang University (Science Edition), 2015, 42(3): 339−346. [45] Wheeler A P, George J W, Evans C A. Control of calcium carbonate nucleation and crystal growth by soluble matrx of oyster shell[J]. Science, 1981, 212(4501): 1397−1398. doi: 10.1126/science.212.4501.1397 [46] 李兴霞. 长牡蛎左右外套膜的分子差异研究[D]. 沈阳: 沈阳农业大学, 2017.Li Xingxia. Study on the molecular difference of the mantle of pacific oyster Crassostrea gigas[D]. Shenyang: Shenyang Agricultural University, 2017. [47] Yan Zhenguang, Jing Gu, Gong Ningping, et al. N40, a novel nonacidic matrix protein from pearl oyster nacre, facilitates nucleation of aragonite in vitro[J]. Biomacromolecules, 2007, 8(11): 3597−3601. doi: 10.1021/bm0701494 [48] Fang Dong, Pan Cong, Lin Huijuan, et al. Novel basic protein, PfN23, functions as key macromolecule during nacre formation[J]. Journal of Biological Chemistry, 2012, 287(19): 15776−15785. doi: 10.1074/jbc.M112.341594 [49] 孔晶晶. 基质蛋白调控晶体自组装形成贝壳有序结构的机制研究[D]. 北京: 清华大学, 2021.Kong Jingjing. Studies on the mechanism of matrix proteins regulating crystals self-assembly to form ordered shell structure[D]. Beijing: Tsinghua University, 2021. [50] Fang Zi, Yan Zhenguang, Li Shuo, et al. Localization of calmodulin and calmodulin-like protein and their functions in biomineralization in P. fucata[J]. Progress in Natural Science, 2008, 18(4): 405−412. doi: 10.1016/j.pnsc.2007.11.011 [51] 于旭蓉, 仇雪梅, 常亚青, 等. 长牡蛎钙调蛋白基因克隆及多态性分析[J]. 南方农业学报, 2012, 43(6): 855−860. doi: 10.3969/j:issn.2095-1191.2012.06.855Yu Xurong, Qiu Xuemei, Chang Yaqing, et al. Cloning and polymorphism analysis of calmodulin (CaM) gene obtained from Crassostrea gigas[J]. Journal of Southern Agriculture, 2012, 43(6): 855−860. doi: 10.3969/j:issn.2095-1191.2012.06.855 [52] Xin Xiaoyu, Liu Chang, Liu Zhaoqu, et al. Calmodulin regulates the calcium homeostasis in mantle of Crassostrea gigas under ocean acidification[J]. Frontiers in Marine Science, 2022, 9: 1050022. doi: 10.3389/fmars.2022.1050022 -




 下载:
下载: