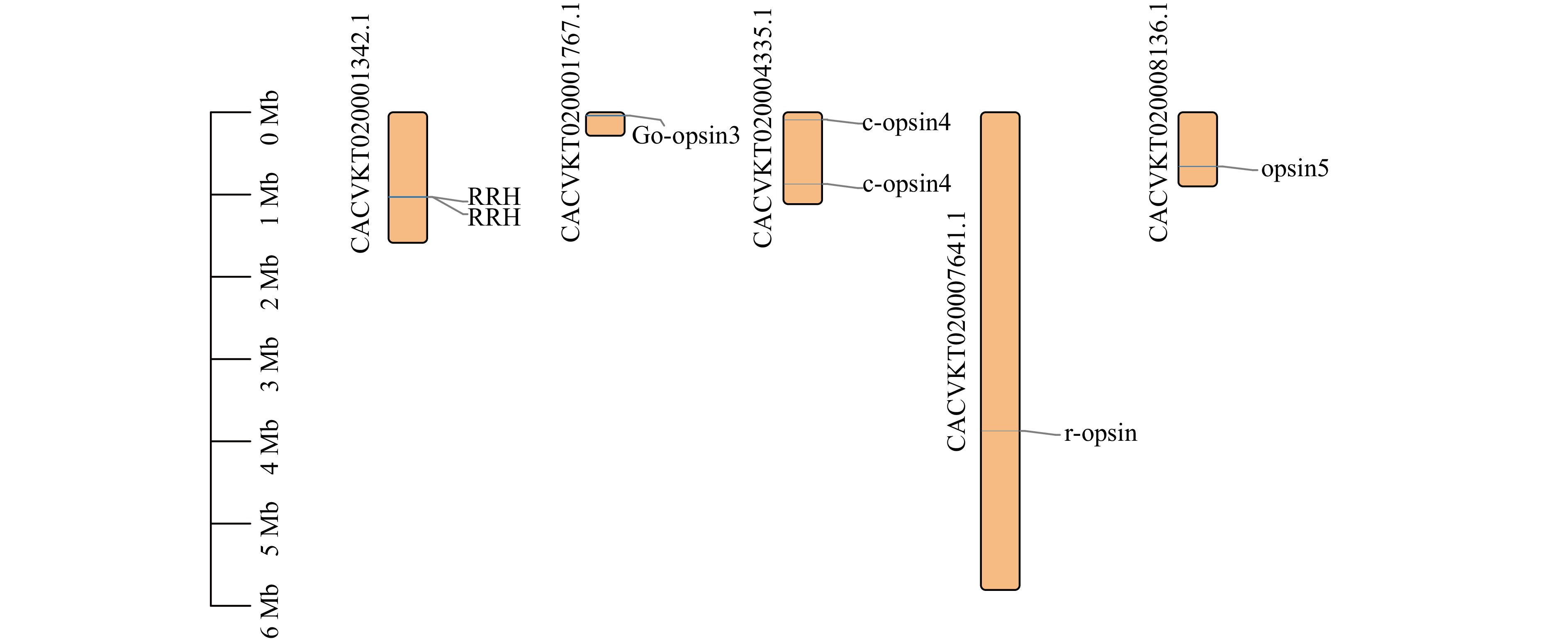
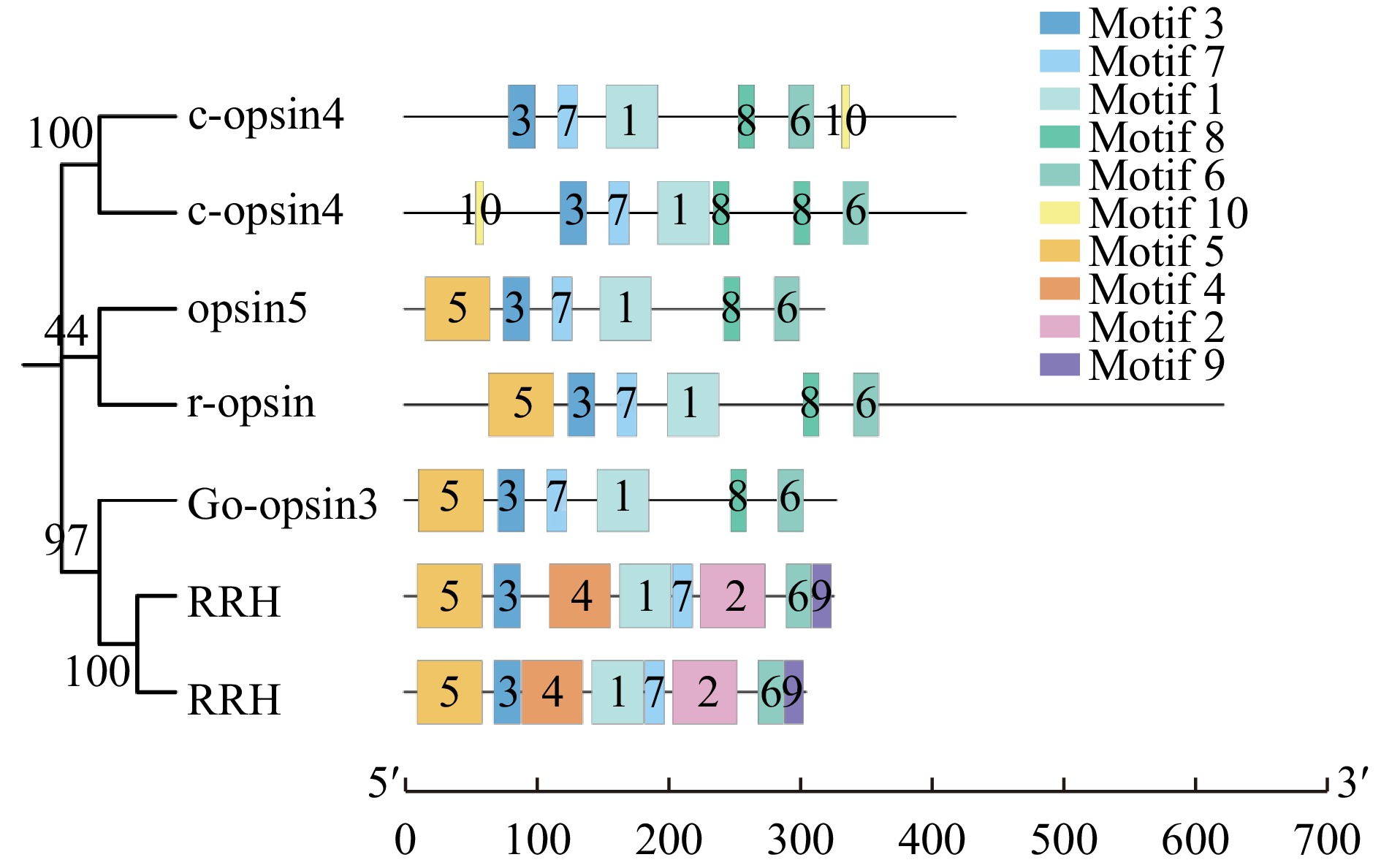
Analysis of opsin gene expression in Mytilus coruscus
-
摘要: 厚壳贻贝是我国重要的经济养殖贝类,光环境可以影响其生活和行为,但其视蛋白基因在很大程度上是未知的。本研究基于厚壳贻贝全基因组数据,共鉴定出7个视蛋白,序列分析结果将其分为5类(r-opsin、c-opsin、Go-opsin、neuropsin和peropsins)。染色体定位结果显示同一亚家族的视蛋白基因位于同一染色体上。生物信息学结果显示,除r-opsin为亲水蛋白,其他蛋白质的均为疏水蛋白。保守基序结果显示视蛋白同一亚家族的视蛋白具有高度的保守性,同时也存在一定的差异。通过实时荧光定量PCR检测厚壳贻贝5个发育时期的视蛋白基因表达量变化。结果显示,不同发育时期的视蛋白基因表达量存在显著差异,c-opsin4和r-opsin在眼点期的表达量显著高于其他时期,表明这两个视蛋白在厚壳贻贝的眼点期中发挥了重要作用。本研究分析了厚壳贻贝视蛋白的分子特征,并初步探究厚壳贻贝发育过程中视蛋白基因的变化规律,为研究厚壳贻贝的视觉形成机制并对视蛋白调控厚壳贻贝变态过程关键分子机制提供科学依据。Abstract: Mytilus coruscus is an economically significant shellfish cultivated in China, with its life and behavior greatly influenced by light conditions. However, much remains unknown about its opsin genes. In this study, seven opsins were identified based on whole-genome sequencing data of M. coruscus, and sequence analysis classified them into five types: r-opsin, c-opsin, Go-opsin, neuropsin, and peropsin. Chromosomal localization analysis revealed that opsin genes of the same subfamily are located on the same chromosome. Bioinformatics analysis showed that, except for r-opsin, all identified proteins are hydrophobic. The conserved motifs revealed high sequence conservation among opsin subfamily members, while inter-subfamily comparisons identified specific divergent residues. The expression profiles of opsin genes were examined across five developmental stages using real-time quantitative PCR, which demonstrated significant expression differences at various developmental stages. Notably, c-opsin4 and r-opsin were significantly upregulated during the eyespot stage, suggesting their crucial roles during this period.This study provides insights into the molecular characteristics of opsins in M. coruscus and preliminarily explores the expression patterns of opsin genes during its development. Additionally, it contributes to the understanding of visual formation mechanisms in M. coruscus and offers a scientific basis for further exploration of the regulatory role of opsins in the metamorphosis process.
-
Key words:
- Mytilus coruscus /
- XXXX /
- opsin /
- expression analysis /
- qPCR
-
图 1 视蛋白基因多序列比对图
红色字体和蓝色字体分别代表本文鉴定的厚壳贻贝和深海偏顶蛤视蛋白序列。图中灰色虚线代表视紫红质的第Ⅲ、Ⅳ和Ⅶ跨膜区。依据牛视紫红质对其他物种中opsin序列的氨基酸位置进行编号,我们可以在其中观察到一些被用于区分opsin家族的保守的氨基酸序列位置,这些位置用红色三角箭头表示。同时,蓝色方框中还标注了r-opsin和c-opsin的特征序列“HPK”和“NKQ”
Fig. 1 Multiple sequence alignment of opsin proteins
The red and blue fonts represent the opsin sequences identified in this paper, respectively. The dashed gray lines in the figure represent the transmembrane regions Ⅲ, Ⅳ, and Ⅶ of the rhodopsin. In the numbering of amino acid positions of opsin sequences in other species by rhodopsin, we can observe some conserved amino acid sequence positions that are used to distinguish the opsin family, and these positions are indicated by red triangular arrows. At the same time, the feature sequences “HPK” and “NKQ” of r-opsin and c-opsin are also marked in the blue box
图 2 视蛋白家族ML系统发育分析
厚壳贻贝的视蛋白序列名用橙色星号标出,深海偏顶蛤的视蛋白序列名用蓝色圆形标出。不同颜色的分枝代表不同的亚家族,树枝上的数字表示ML后验概率
Fig. 2 Phylogenetic relationship of opsin family based on a ML analysis of protein sequences
The opsin sequence names of Mytilus coruscus are marked with orange asterisks, and those of Bathymodiolus platifrons are marked with blue circles. Different colored branches represent different subfamilies, and the numbers on the branches represent ML posteriori probabilities
表 1 视蛋白家族序列及其NCBI登录号
Tab. 1 Sequence of opsin family and their NCBI accession number
物种名 蛋白 登录号 物种名 蛋白 登录号 文昌鱼 Branchiostoma belcheri opsin6 BAC76024 光滑双脐螺 Biomphalaria glabrata pineal opsin like XP_055862616 melanopsin Q4R114 opsin5 like XP_055889746 opsin2 BAC76020 葡萄牙牡蛎 Crassostrea angulata opsin VA like XP_052683732 peropsin DAC74078 pineal opsin like XP_052683447 opsin1 BAC76019 戴河笠贝 Lottia peitaihoensis r-opsin WKW95581 opsin3 BAC76023 neurospin WKW95586 opsin5 BAC76022 Go-opsin3 WKW95588 opsin4 BAC76021 peropsin WKW95585 neuropsin DAC74071 虎斑乌贼 Sepia pharaonis RRH CAE1253240 海湾扇贝 Argopecten irradians opsin Gq2 ALO02515 opsin4 CAE1318583 opsin Gq3 ALO02516 地中海贻贝 Mytilusgalloprovincialis r-opsin VDI13980 opsin Gq4 ALO02517 虾夷扇贝 Mizuhopecten yessoensis Go-coupled opsin XP_021355729 opsin Gq1 ALO02514 日本大鲍 Haliotis gigantea rhodopsin UVT38273 Rhabdomeric opsin AUR34014 欧洲扇贝 Pecten maximus retinochrome UUS63783 Go-coupled opsin AUR34012 夏威夷短尾乌贼 Euprymna scolopes opsin ACB05673 Go-coupled opsin1 APB88014 乌贼 Sepia officinalis rhodopsin ADT78509 人 Homo sapiens encephalopsin NP055137 尖膀胱螺 Physella acuta opsin 3 like XP_059165397 opsin3 AAH36773 硬壳蛤 Mercenaria mercenaria opsin 5 like XP_045202610 blue opsin P0399 砂海螂 Mya arenaria opsin 5 like XP_052784401 rhodopsin NP000530 Ambigolimax valentianus rhodopsin BBA21101 green opsin P04001 Ambigolimax valentianus opsin 5B BBH84662 red opsin P04000 Acanthopleura granulata r-opsin r-opsin melanopsin NP150598 Ambigolimax valentianus retinochrome BBH84663 opsin5 QU6736 Chiton virgulatus opsin APF30602 peropsin NP006574 Chiton tuberculatus r-opsin QKY89064 鼠 Mus musculus melanopsin AAF24979 Chiton marmoratus r-opsin QKY89065 peropsin AAC53344 Chaetopleura apiculata r-opsin QKY89066 opsin5 NP861418 Elysia marginata rhodopsin GFS12646 褐家鼠 Rattus norvegicus opsin5 NP861437 Leptochiton asellus xenopsin ASM47594 厚壳贻贝 Mytilus coruscus opsin3 CAC5413267 Stenoplax conspicua opsin APF30603 opsin5 CAC5413461 Ylistrum balloti opsin 5 like XP_060063722 紫贻贝 Mytilus edulis opsin5 CAG2190128 RRH CAG2194598 opsin4 CAG2193259 表 2 本实验所用的引物序列
Tab. 2 Primer sequences used in this study
蛋白 引物序列(5'-3') c-opsin4 CATTAGTATGGGCGGTTCCTC TCGTGTTTCCTTATTGCTTTCC r-opsin TGAAAACCCCGAGCAACAT CATCGCCCCACATCCATT Go-opsin3 TAGGAAGGTAACATTGGCGTCTC GCTGCTCTTGGCAAACATCG opsin5 CAAACGCCGAGATGCATAA TGAACTGATACTATTAGAACACGGA RRH ATGCGGACTAAACTGGCTAAAA CTCCTGACCACGGGTAACAAC EF1a CACCACGAGTCTCTCCCTGA GCTGTCACCACAGACCATTCC 表 3 厚壳贻贝视蛋白家族序列基本信息
Tab. 3 Basic information of opsin family members in Mytilus coruscus
基因 氨基酸数目 等电点/PI 分子量/kDa 不稳定性指数 脂肪族指数 GRAVY指数 c-opsin4 416 9.05 46.18 45.90 101.42 0.316 c-opsin4 424 9.21 47.30 442.81 99.93 0.341 r-opsin 619 8.39 69.22 40.88 83.94 -0.145 Go-opsin3 326 9.05 37.22 29.62 114.42 0.165 opsin5 317 9.44 35.98 42.10 117.98 0.702 RRH 324 7.46 36.48 37.02 99.85 0.527 RRH 303 6.96 33.83 33.27 99.70 0.525 -
[1] Terakita A. The opsins[J]. Genome Biology, 2005, 6(3): 213. doi: 10.1186/gb-2005-6-3-213 [2] Cortesi F, Musilová Z, Stieb S M, et al. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(5): 1493−1498. [3] Porter M L, Blasic J R, Bok M J, et al. Shedding new light on opsin evolution[J]. Proceedingsof the Royal Society B: Biological Sciences, 2012, 279(1726): 3−14. doi: 10.1098/rspb.2011.1819 [4] Ramirez M D, Pairett A N, Pankey M S, et al. The last common ancestor of most bilaterian animals possessed at least nine opsins[J]. Genome Biology and Evolution, 2016, 8(12): 3640−3652. [5] Döring C C, Kumar S, Tumu S C, et al. The visual pigment xenopsin is widespread in protostome eyes and impacts the view on eye evolution[J]. Elife, 2020, 9: e55193. doi: 10.7554/eLife.55193 [6] Sato K, Yamashita T, Haruki Y, et al. Two UV-sensitive photoreceptor proteins, Opn5m and Opn5m2 in ray-finned fish with distinct molecular properties and broad distribution in the retina and brain[J]. PLoS One, 2016, 11(5): e0155339. doi: 10.1371/journal.pone.0155339 [7] Beaudry F E G, Iwanicki T W, Mariluz B R Z, et al. The non-visual opsins: eighteen in the ancestor of vertebrates, astonishing increase in ray-finned fish, and loss in amniotes[J]. Journal of Experiment al Zoology Part B, Molecular and Developmental Evolution, 2017, 328(7): 685−696. doi: 10.1002/jez.b.22773 [8] 任红乐. 花鲈视蛋白家族分析及OPN5对光周期的响应[D]. 上海: 上海海洋大学, 2023.Ren Hongle. Analysis of opsin gene family and response of OPN5 to photoperiod of Lateolabrax maculatus[D]. Shanghai: Shanghai Ocean University, 2023. [9] 张志博, 吴燕玲, 张怡宁, 等. 绿光辐照度对红鳍东方鲀视网膜、视蛋白基因及氧化应激的影响[J]. 渔业现代化, 2024, 51(3): 8−16. doi: 10.3969/j.issn.1007-9580.2024.03.002Zhang Zhibo, Wu Yanling, Zhang Yining, et al. Effects of green light irradiance on retinal, opsins genes and oxidative stress of Takifugu rubripes[J]. Fishery Modernization, 2024, 51(3): 8−16. doi: 10.3969/j.issn.1007-9580.2024.03.002 [10] 刘峰, 吴乐乐, 王雨浓, 等. 大菱鲆幼鱼视网膜结构及不同光谱下视蛋白基因表达特征的研究[J]. 海洋科学, 2023, 47(12): 30−39.Liu Feng, Wu Lele, Wang Yunong, et al. Development of retinal structures and opsin gene expression under different spectra in juvenile turbot[J]. Marine Sciences, 2023, 47(12): 30−39. [11] Bellingham J, Morris A G, Hunt D M. The rhodopsin gene of the cuttlefish Sepia officinalis: sequence and spectral tuning[J]. Journal of Experimental Biolgy, 1998, 201(15): 2299−2306. doi: 10.1242/jeb.201.15.2299 [12] Ovchinnikov Y A, Abdulaev N G, Zolotarev A S, et al. Octopus rhodopsin amino acid sequence deduced from cDNA[J]. FEBS Letters, 1988, 232(1): 69−72. doi: 10.1016/0014-5793(88)80388-0 [13] Katagiri N, Terakita A, Shichida Y, et al. Demonstration of a rhodopsin-retinochrome system in the stalk eye of a marine gastropod, Onchidium, by immunohistochemistry[J]. Journal of Comparative Neurology, 2001, 433(3): 380−389. doi: 10.1002/cne.1146 [14] Donohue M W, Carleton K L, Cronin T W. Opsin expression in the central nervous system of the mantis shrimp Neogonodactylus oerstedii[J]. The Biological Bulletin, 2017, 233(1): 58−69. doi: 10.1086/694421 [15] Levy O, Appelbaum L, Leggat W, et al. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora[J]. Science, 2007, 318(5849): 467−470. doi: 10.1126/science.1145432 [16] Fritzenwanker J H, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa)[J]. Development Genes and Evolution, 2002, 2,212(2): 99−103. [17] Macias-Muñoz A, Murad R, Mortazavi A. Molecular evolution and expression of opsin genes in Hydra vulgaris[J]. BMC Genomics, 2019, 20(1): 992. doi: 10.1186/s12864-019-6349-y [18] Ullrich-Lüter E M, Dupont S, Arboleda E, et al. Unique system of photoreceptors in sea urchin tube feet[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(20): 8367−8372. [19] Delroisse J, Lanterbecq D, Eeckhaut I, et al. Opsin detection in the sea urchin Paracentrotus lividus and the sea star Asterias rubens[J]. Cahiers de Biologie Marine, 2013, 54(4): 721−727. [20] Kong Fei, Ran Zhaoshou, Zhang Mengqi, et al. Eyeless razor clam Sinonovacula constricta discriminates light spectra through opsins to guide Ca2+ and cAMP signaling pathways[J]. Journal of Biological Chemistry, 2024, 300(1): 105527. doi: 10.1016/j.jbc.2023.105527 [21] Hasan M S, McElroy K E, Audino J A, et al. Opsin expression varies across larval development and taxa in pteriomorphian bivalves[J]. Frontiers in Neuroscience 2024, 18: 1357873. [22] Niemiller M L, Fitzpatrick B M, Shah P, et al. Evidence for repeated loss of selective constraint in rhodopsin of amblyopsid cavefishes (teleostei: amblyopsidae)[J]. Evolution, 2013, 67(3): 732−748. doi: 10.1111/j.1558-5646.2012.01822.x [23] McElroy K E, Audino J A, Serb J M. Molluscan genomes reveal extensive differences in photopigment evolution across the phylum[J]. Molecular Biology and Evolution, 2023, 40(12): msad263. doi: 10.1093/molbev/msad263 [24] Koehn R K. The genetics and taxonomy of species in the genus Mytilus[J]. Aquaculture, 1991, 94(2/3): 125−145. [25] Li Ronghua, Zhang Weijia, Lu Junkai, et al. The whole-genome sequencing and hybrid assembly of Mytilus coruscus[J]. Frontiers in Genetics, 2020, 11: 440. doi: 10.3389/fgene.2020.00440 [26] Yan Chengrui, Xu Minhui, Ye Yingying, et al. Hox gene clusters in the mussel Mytilus coruscus: Implications for bivalves’ evolution[J]. Ecological Indicators, 2023, 154: 110810. doi: 10.1016/j.ecolind.2023.110810 [27] Chen Chengjie, Chen Hao, Zhang Yi, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194−1202. doi: 10.1016/j.molp.2020.06.009 [28] Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the $ 2^{-\Delta \Delta C_{T}} $ method[J]Methods, 2001, 25(4): 402−408. [29] Say T E, Degnan S M. Molecular and behavioural evidence that interdependent photo-and chemosensory systems regulate larval settlement in a marine sponge[J]. Molecular Ecology, 2020, 29(2): 247−261. doi: 10.1111/mec.15318 [30] 孙妍. 扇贝眼睛发生和视觉功能遗传基础的基因组学研究[D]. 青岛: 中国海洋大学, 2016.Sun Yan. Genomic study on genetic basis of eye development and visual function in scallop[D]. Qingdao: Ocean University of China, 2016. [31] De Vivo G, Crocetta F, Ferretti M, et al. Duplication and losses of opsin genes in lophotrochozoan evolution[J]. Molecular Biology and Evolution, 2023, 40(4): msad066. doi: 10.1093/molbev/msad066 [32] Carl C, Poole A J, Williams M R, et al. Where to settle—settlement preferences of Mytilus galloprovincialis and choice of habitat at a micro spatial scale[J]. PLoS One, 2012, 7(12): e52358. doi: 10.1371/journal.pone.0052358 [33] Vöcking O, Kourtesis I, Tumu S C, et al. Co-expression of xenopsin and rhabdomeric opsin in photoreceptors bearing microvilli and cilia[J]. ELife, 2017, 6: e23435. doi: 10.7554/eLife.23435 [34] Rawlinson K A, Lapraz F, Ballister E R, et al. Extraocular, rod-like photoreceptors in a flatworm express xenopsin photopigment[J]. ELife, 2019, 8: e45465. doi: 10.7554/eLife.45465 [35] Randel N, Bezares-Calderón L A, Gühmann M, et al. Expression dynamics and protein localization of rhabdomeric opsins in Platynereis larvae[J]. Integrative and Comparative Biology, 2013, 53(1): 7−16. doi: 10.1093/icb/ict046 [36] Hodgson C A, Burke R D. Development and larval morphology of the spiny scallop, Chlamys hastata[J]. The Biological Bulletin, 1988, 174(3): 303−318. doi: 10.2307/1541956 [37] Lorion J, Kiel S, Faure B, et al. Adaptive radiation of chemosymbiotic deep-sea mussels[J]. Proceedings of the Royal Society B: Biological Sciences, 2013, 280(1770): 20131243. doi: 10.1098/rspb.2013.1243 [38] Mat A M, Sarrazin J, Markov G V, et al. Biological rhythms in the deep-sea hydrothermal mussel Bathymodiolus azoricus[J]. Nature Communications, 2020, 11(1): 3454. doi: 10.1038/s41467-020-17284-4 [39] Shen Weiping, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression[J]. Development, 1997, 124(1): 45−52. doi: 10.1242/dev.124.1.45 [40] Senthilan P R, Piepenbrock D, Ovezmyradov G, et al. Drosophila auditory organ genes and genetic hearing defects[J]. Cell, 2012, 150(5): 1042−1054. doi: 10.1016/j.cell.2012.06.043 [41] Lythgoe J N. The Ecology of Vision[M]. New York: Clarendon Press, 1979. [42] Bybee S M, Johnson K K, Gering E J, et al. All the better to see you with: a review of odonate color vision with transcriptomic insight into the odonate eye[J]. Organisms Diversity & Evolution, 2012, 12(3): 241−250. [43] Sun Hui, Gilbert D J, Copeland N G, et al. Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(18): 9893−9898. [44] Hagen J F D, Roberts N S, Johnston Jr R J. The evolutionary history and spectral tuning of vertebrate visual opsins[J]. Developmental Biology, 2023, 493: 40−66. doi: 10.1016/j.ydbio.2022.10.014 -




 下载:
下载: