Changes of microbial community composition in porites lutea during health-bleaching recovery under high temperature stress
-
摘要: 热胁迫下珊瑚白化现象已被认为是全球珊瑚礁退化的主要驱动因素。热胁迫条件下珊瑚全共生功能体内的微生物群落组成、代谢和功能特性已有相关研究报道。然而,迄今还未见珊瑚健康-白化-恢复全过程中菌群结构及组成变化的相关研究。本研究以深圳海域橙黄滨珊瑚(Porites lutea)为研究对象,通过实验室模拟热胁迫下的珊瑚从健康-白化-恢复健康的过程,采用高通量测序技术和宏基因组测序技术分析珊瑚白化和恢复过程中的健康、开始白化、持续白化、开始恢复、恢复健康5个特征阶段珊瑚微生物群落的差异和功能基因变化。结果显示,随着温度的升高,变形杆菌门在白化过程中明显增多,在恢复过程中减少;拟杆菌门等在白化过程中减少,在恢复过程中增多。在珊瑚白化过程中,与压力耐受能力、生物膜形成、可移动因子及潜在致病性相关的细菌丰度显著增加,而群体感应相关细菌的丰度则在减少。4种微生物在珊瑚白化中发挥重要作用:不动杆菌属、罗尔斯通菌属和伯克霍尔德氏菌属是升温导致的珊瑚白化中的关键差异细菌类群;代尔夫特菌属可能通过群体感应机制调节其他菌群,以维持珊瑚微生物群落的稳定。本研究揭示了高温胁迫环境下珊瑚组织内微生物及其功能的变化,为阐明珊瑚白化过程中微生物−宿主互作机制提供分子依据。Abstract: Global coral bleaching under heat stress has been identified as a major driver of coral reef degradation. The composition, metabolism and functional characteristics of microbial communities in coral holosomes under heat stress has been reported. However, the changes in microbial structure and composition throughout the entire process of coral health−bleaching−recovery have not been studied so far. In this study, Porites lutea in Shenzhen Sea area was selected as the research object. The process of coral health−bleaching−recovery under heat stress was simulated in the laboratory. High-throughput sequencing and macro-genome sequencing technologies were used to analyse the differences in coral microbial communities and functional gene changes during five characteristic phases of coral bleaching and restoration: healthy, beginning of bleaching, continued bleaching, beginning of recover, and recovered. With the increase of temperature, Proteobacteria increased significantly during the bleaching process and decreased during recovery; Bacteroidota etc. decreased during bleaching and increased during recovery. During the coral bleaching process, the abundance of bacteria associated with stress tolerance, biofilm formation, mobile elements, and potential pathogenicity significantly increases. Conversely, the abundance of bacteria involved in quorum sensing decreases. Notably,four kinds of microbes play a crucial role in coral bleaching: Acinetobacter, Rhodobacter, and Burkholderia are key differential taxa in warming-induced coral bleaching, while Delftia may modulate other bacterial assemblages via quorum sensing mechanisms to maintain the stability of coral microbial communities.This study revealed the changes of microbes and their functions in coral tissues under high temperature stress, which provided molecular basis for elucidating the interaction mechanism between microbes and hosts during coral bleaching.
-
Key words:
- coral bleaching /
- pure bacteria /
- high-throughput sequencing /
- metagenome
-
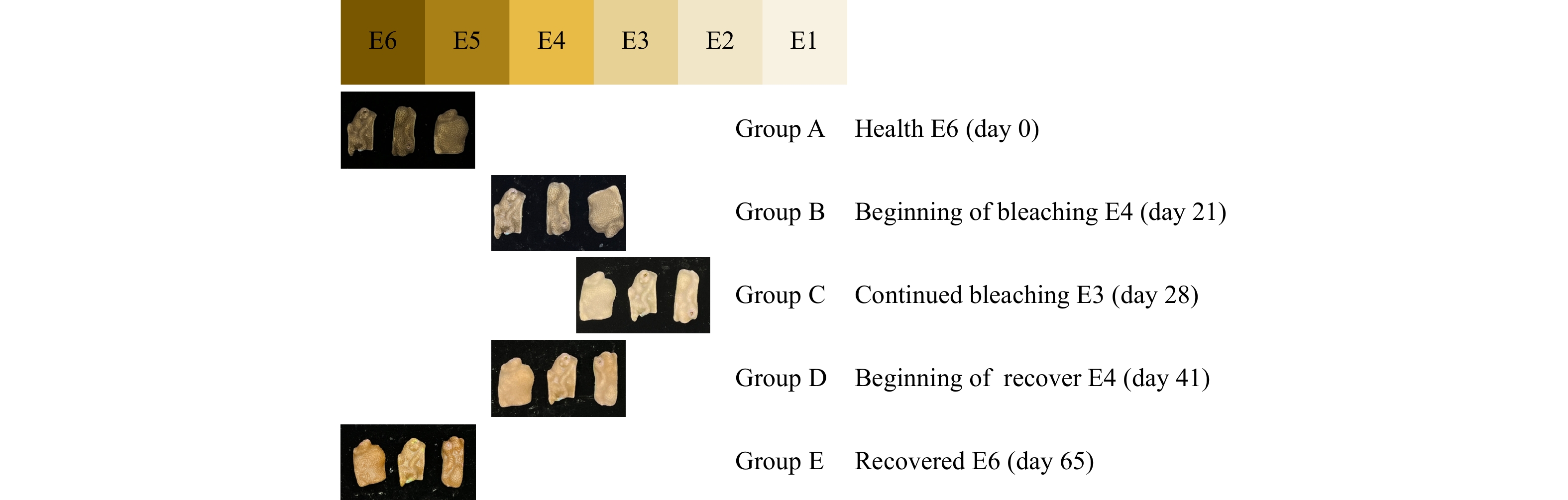
图 2 微生物群落α多样性指数图
a−d分别代表Shannon、Coverage、Simpson和Chao指数。颜色代表不同分组。健康 E6(A组)、开始白化E4(B组) 、持续白化 E3(C组)、开始恢复E4(D组)、恢复健康E6(E组),下同
Fig. 2 Plot of alpha diversity index of microbial community
a-d represents shannon,coverage,simpson and chao indices,respectively. The colors represent the different groups. Healthy E6 (group A), beginning of bleaching E4(group B), continued bleaching E3 (group C), beginning of recover E4(group D), recovered E6 (group E), the same below
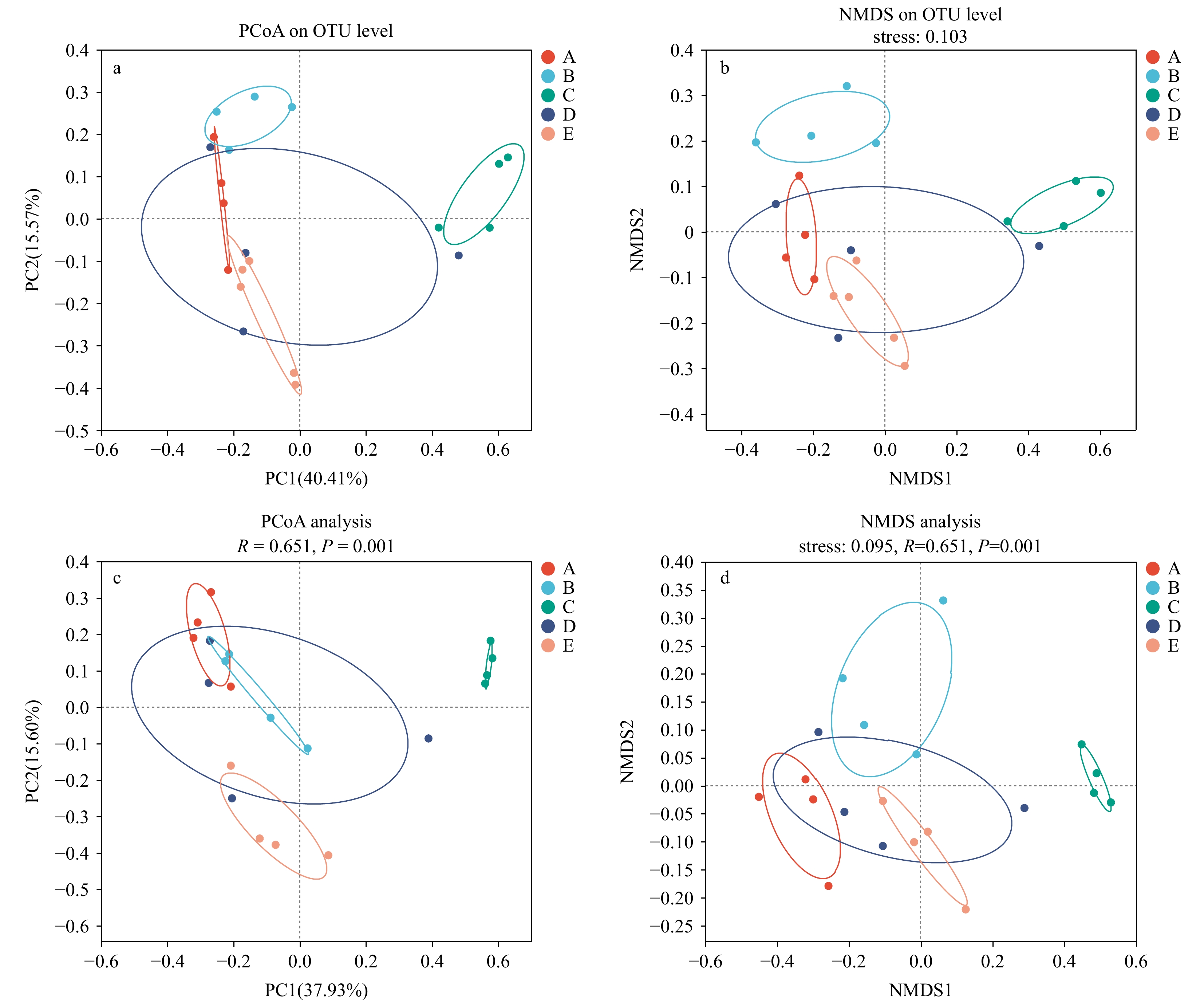
图 4 16S rRNA高通量测序结果在OTU水平上的微生物群落结构主坐标分析(PCoA)(a)和非度量多维尺度分析(NMDS)(b),宏基因组测序结果在物种水平上的微生物群落结构主坐标分析(c)与非度量多维尺度分析(d),颜色代表不同分组
Fig. 4 Microbial community structure principal coordinate analysis (PCoA)(a) and non-metric multidimensional scaling (NMDS)(b) of 16S rRNA high-throughput sequencing results at the OTU level, and microbial community structure principal coordinate analysis (c) and non-metric multidimensional scaling (d) of metagenome sequencing results at the species level. Colors represent different groups
图 5 基于16S rRNA高通量测序的珊瑚组织中细菌群落组成结构与差异
a,门水平下珊瑚组织中细菌群落组成;b,属水平下珊瑚组织中细菌群落组成
Fig. 5 Composition and diversity of bacterial communities in coral tissues based on 16S rRNA high-throughput sequencing
a,Bacterial community composition in coral tissues at phylum level; b,bacterial community composition in coral tissues at genus level
图 6 基于BugBase数据库的属水平上细菌群落表型预测
每组中5个平行样合并后进行分析。a−c,好氧、兼性厌氧和厌氧细菌。d−e,革兰氏阴性菌和革兰氏阳性菌。f,潜在致病菌。g−i. 与压力耐受、生物膜形成、可移动因子相关的细菌。图中展示了丰度最高的5个属
Fig. 6 Prediction of bacterial community phenotypes at the genus level based on BugBase database
Five parallel samples from each group were pooled and analyzed. a−c. aerobic, facultative anaerobic, and anaerobic bacteria. d−e. Gram-negative and Gram-positive bacteria. f. Potential pathogenic bacteria. g−i. bacteria associated with pressure tolerance, biofilm formation, and mobile factors. The five most abundant genera are shown
图 7 基于宏基因组测序的珊瑚白化和恢复过程中珊瑚组织中细菌群落门水平上的差异
a,门水平下珊瑚组织中细菌群落组成;b,属水平下珊瑚组织中细菌群落组成
Fig. 7 Metagenomic sequencing results: differences in the phyla level of bacterial communities in coral tissues during coral bleaching and recovery
a, Composition of bacterial community in coral tissue at portal level; b, composition of bacterial community in coral tissue below genus level
图 8 高温胁迫前后5组珊瑚共生菌群落属水平上的共线性网络图
图中选取丰度前50的属进行分析。a、c、e、g和i分别为A−E组;b、d、f、h和j分别为对应组中与不动杆菌属、伯克霍尔德菌属、代尔夫特菌属和罗尔斯通菌属有关联的细菌属相关性网络图。图中圆点的颜色代表不同细菌门,圆点之间的线代表相关性。红色线代表正相关,绿色线代表负相关
Fig. 8 Collinear network plots at the genus level of five coral commensal communities before and after high temperature stress
The top 50 abundance genera in the figure were selected for analysis. a, c, e, g and i were groups A−E, respectively. b, d, f, h, and j show the bacterial genus correlation networks associated with Acinetobacter, Burkholderia, Delftia, and Ralstonia in the corresponding group, respectively. The colors of the dots in the figure represent the different bacterial phyla, and the lines between the dots represent correlations. The red line represents a positive correlation and the green line represents a negative correlation
表 1 各组序列信息均值表
Tab. 1 Table of mean values of sequence information for each group
组 OTU num 序列/条 A 2118 48344 B 2415 48344 C 794 48344 D 1997 48344 E 1965 48344 表 2 珊瑚内微生物宏基因组测序结果
Tab. 2 Results of metagenomic sequencing of microorganisms in coral
Sample Raw reads Clean reads Clean Q30/% Clean GC content/% A1 125890046 122724428 95.90 41.19 A2 82659748 80544214 95.71 40.70 A3 89049322 86754636 95.80 40.66 A4 106771568 104045876 95.78 43.31 B1 90936840 89390946 92.96 41.55 B2 89245844 85449436 93.54 40.19 B3 91917958 88081606 93.69 41.19 B4 86496526 85024148 93.62 41.75 C1 92341038 89439794 93.56 39.73 C2 91189792 88130800 93.43 41.45 C3 91013390 87703802 92.98 43.40 C4 105006296 100526562 93.60 41.30 D1 87205816 85689258 93.22 41.42 D2 86749008 84530530 95.64 40.16 D3 84502268 82332052 95.73 40.78 D4 88563536 87162758 92.78 41.66 E1 82887590 81573156 93.53 41.55 E2 86973042 85542298 93.27 41.55 E3 86551346 84987090 93.06 41.59 E4 85688114 84203114 93.02 41.05 -
[1] Polidoro B, Carpenter K. Dynamics of coral reef recovery[J]. Science, 2013, 340(6128): 34−35. doi: 10.1126/science.1236833 [2] Doney S C, Ruckelshaus M, Emmett Duffy J, et al. Climate change impacts on marine ecosystems[J]. Annual Review of Marine Science, 2012, 4(1): 11−37. doi: 10.1146/annurev-marine-041911-111611 [3] Manikandan B, Ravindran J, Shrinivaasu S, et al. Community structure and coral status across reef fishing intensity gradients in Palk Bay reef, southeast coast of India[J]. Environmental Monitoring and Assessment, 2014, 186(10): 5989−6002. doi: 10.1007/s10661-014-3835-1 [4] Hughes T P, Kerry J T, Álvarez-Noriega M, et al. Global warming and recurrent mass bleaching of corals[J]. Nature, 2017, 543(7645): 373−377. doi: 10.1038/nature21707 [5] Frölicher T L, Fischer E M, Gruber N. Marine heatwaves under global warming[J]. Nature, 2018, 560(7718): 360−364. doi: 10.1038/s41586-018-0383-9 [6] Chen Tianrun, Yu Kefu, Shi Qi, et al. Twenty-five years of change in scleractinian coral communities of Daya Bay (northern South China Sea) and its response to the 2008 AD extreme cold climate event[J]. Chinese Science Bulletin, 2009, 54(12): 2107−2117. doi: 10.1007/s11434-009-0007-8 [7] Yu Kefu. Coral reefs in the South China Sea: their response to and records on past environmental changes[J]. Science China Earth Sciences, 2012, 55(8): 1217−1229. doi: 10.1007/s11430-012-4449-5 [8] Charpy L, Casareto B E, Langlade M J, et al. Cyanobacteria in coral reef ecosystems: a review[J]. Journal of Marine Sciences, 2012, 2012(1): 259571. [9] Tout J, Jeffries T C, Webster N S, et al. Variability in microbial community composition and function between different niches within a coral reef[J]. Microbial Ecology, 2014, 67(3): 540−552. doi: 10.1007/s00248-013-0362-5 [10] Glasl B, Webster N S, Bourne D G. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems[J]. Marine Biology, 2017, 164(4): 91. doi: 10.1007/s00227-017-3097-x [11] Shade A, Peter H, Allison S D, et al. Fundamentals of microbial community resistance and resilience[J]. Frontiers in Microbiology, 2012, 3: 417. [12] Liu Zhuhong, Chen Chang, Gao Lei, et al. Differences in microbial communities between healthy and bleached coral Acropora solitaryensis from Xisha Islands, South China Sea[J]. Marine Biology Research, 2016, 12(10): 1101−1108. doi: 10.1080/17451000.2016.1236201 [13] 周进, 晋慧, 蔡中华. 微生物在珊瑚礁生态系统中的作用与功能[J]. 应用生态学报, 2014, 25(3): 919−930.Zhou Jin, Jin Hui, Cai Zhonghua. A review of the role and function of microbes in coral reef ecosystem[J]. Chinese Journal of Applied Ecology, 2014, 25(3): 919−930. [14] Ziegler M, Seneca F O, Yum L K, et al. Bacterial community dynamics are linked to patterns of coral heat tolerance[J]. Nature Communications, 2017, 8(1): 14213. doi: 10.1038/ncomms14213 [15] Qin Zhenjun, Yu Kefu, Liang Jiayuan, et al. Significant changes in microbial communities associated with reef corals in the southern South China Sea during the 2015/2016 global‐scale coral bleaching event[J]. Journal of Geophysical Research: Oceans, 2020, 125(7): e2019JC015579. doi: 10.1029/2019JC015579 [16] Xie J Y, Yeung Y H, Kwok C K, et al. Localized bleaching and quick recovery in Hong Kong's coral communities[J]. Marine Pollution Bulletin, 2020, 153: 110950. doi: 10.1016/j.marpolbul.2020.110950 [17] Bourne D, Iida Y, Uthicke S, et al. Changes in coral-associated microbial communities during a bleaching event[J]. The ISME Journal, 2008, 2(4): 350−363. doi: 10.1038/ismej.2007.112 [18] Littman R, Willis B L, Bourne D G. Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef[J]. Environmental Microbiology Reports, 2011, 3(6): 651−660. doi: 10.1111/j.1758-2229.2010.00234.x [19] Ben-Haim Y, Rosenberg E. A novel Vibrio sp. pathogen of the coral Pocillopora damicornis[J]. Marine Biology, 2002, 141(1): 47−55. doi: 10.1007/s00227-002-0797-6 [20] Oladi M, Shokri M R, Rajabi-Maham H. Application of the coral health chart to determine bleaching status of Acropora downingi in a subtropical coral reef[J]. Ocean Science Journal, 2017, 52(2): 267−275. doi: 10.1007/s12601-017-0025-4 [21] 黄晖, 杨泽民, 张俊彬, 等. 柳珊瑚4种不同DNA提取方法的比较研究[J]. 海洋通报, 2007, 26(2): 100−104. doi: 10.3969/j.issn.1001-6392.2007.02.015Huang Hui, Yang Zemin, Zhang Junbin, et al. Comparison of 4 different gorgonian DNA extraction methods[J]. Marine Science Bulletin, 2007, 26(2): 100−104. doi: 10.3969/j.issn.1001-6392.2007.02.015 [22] Walters W, Hyde E R, Berg-Lyons D, et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys[J]. Msystems, 2016, 1(1): 10. 1128. [23] Li Dinghua, Liu Chiman, Luo Ruibang, et al. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph[J]. Bioinformatics, 2015, 31(10): 1674−1676. doi: 10.1093/bioinformatics/btv033 [24] Fu Limin, Niu Beifang, Zhu Zhengwei, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150−3152. doi: 10.1093/bioinformatics/bts565 [25] Li Ruiqiang, Yu Chang, Li Yingrui, et al. SOAP2: an improved ultrafast tool for short read alignment[J]. Bioinformatics, 2009, 25(15): 1966−1967. doi: 10.1093/bioinformatics/btp336 [26] Wyatt A S J, Leichter J J, Toth L T, et al. Heat accumulation on coral reefs mitigated by internal waves[J]. Nature Geoscience, 2019, 13(1): 28−34. [27] Gardner S G, Camp E F, Smith D J, et al. Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave[J]. Ecology and Evolution, 2019, 9(3): 938−956. doi: 10.1002/ece3.4662 [28] Ziegler M, Grupstra C G B, Barreto M M, et al. Coral bacterial community structure responds to environmental change in a host-specific manner[J]. Nature Communications, 2019, 10(1): 3092. doi: 10.1038/s41467-019-10969-5 [29] Glasl B, Herndl G J, Frade P R. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance[J]. The ISME Journal, 2016, 10(9): 2280−2292. doi: 10.1038/ismej.2016.9 [30] Suggett D J, Smith D J. Coral bleaching patterns are the outcome of complex biological and environmental networking[J]. Global Change Biology, 2020, 26(1): 68−79. doi: 10.1111/gcb.14871 [31] 林姿君, 蔡中华, 林光辉, 等. 健康与白化状态下珊瑚共生菌的群落组成与功能差异[J]. 海洋学报, 2018, 40(2): 104−116.Lin Zijun, Cai Zhonghua, Lin Guanghui, et al. Community composition and functional differences of symbiotic bacteria in healthy and blenching coral[J]. Haiyang Xuebao, 2018, 40(2): 104−116. [32] Zaneveld J R, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes[J]. Nature Microbiology, 2017, 2(9): 17121. doi: 10.1038/nmicrobiol.2017.121 [33] 徐帅良. 滨珊瑚共附生细菌多样性及其纯培养研究[D]. 南宁: 广西大学, 2020.Xu Shuailiang. Diversity of Porites. associated bacteria and its pure culture study[D]. Nanning: Guangxi University, 2020. [34] Van De Water J A J M, Allemand D, Ferrier-Pagès C. Host-microbe interactions in octocoral holobionts - recent advances and perspectives[J]. Microbiome, 2018, 6(1): 64. doi: 10.1186/s40168-018-0431-6 [35] Chapelle E, Mendes R, Bakker P A H M, et al. Fungal invasion of the rhizosphere microbiome[J]. The ISME Journal, 2016, 10(1): 265−268. doi: 10.1038/ismej.2015.82 [36] Sharp K H, Distel D, Paul V J. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides[J]. The ISME Journal, 2012, 6(4): 790−801. doi: 10.1038/ismej.2011.144 [37] Kusdianto H, Kullapanich C, Palasuk M, et al. Microbiomes of healthy and bleached corals during a 2016 thermal bleaching event in the upper Gulf of Thailand[J]. Frontiers in Marine Science, 2021, 8: 643962. doi: 10.3389/fmars.2021.643962 [38] Du Jikun, Xiao Kai, Huang Yali, et al. Seasonal and spatial diversity of microbial communities in marine sediments of the South China Sea[J]. Antonie Van Leeuwenhoek, 2011, 100(3): 317−331. doi: 10.1007/s10482-011-9587-9 [39] Alexandre A, Laranjo M, Young J P W, et al. DnaJ is a useful phylogenetic marker for alphaproteobacteria[J]. International Journal of Systematic and Evolutionary Microbiology, 2008, 58(12): 2839−2849. doi: 10.1099/ijs.0.2008/001636-0 [40] Brettar I, Christen R, Höfle M G. Rheinheimera perlucida sp. nov. , a marine bacterium of the Gammaproteobacteria isolated from surface water of the central Baltic Sea[J]. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(9): 2177-2183. [41] Sun Fulin, Wang Youshao, Wu Meilin, et al. Spatial and vertical distribution of bacteria in the Pearl River estuary sediment[J]. African Journal of Biotechnology, 2012, 11(9): 2256−2266. [42] Miura N, Motone K, Takagi T, et al. Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus[J]. Marine Biotechnology, 2019, 21(1): 1−8. doi: 10.1007/s10126-018-9853-1 [43] Kitamura R, Miura N, Ito M, et al. Specific detection of coral-associated Ruegeria, a potential probiotic bacterium, in corals and subtropical seawater[J]. Marine Biotechnology, 2021, 23(4): 576−589. doi: 10.1007/s10126-021-10047-2 [44] Jones P R, Cottrell M T, Kirchman D L, et al. Bacterial community structure of biofilms on artificial surfaces in an estuary[J]. Microbial Ecology, 2007, 53(1): 153−162. doi: 10.1007/s00248-006-9154-5 [45] Cárdenas A, Rodriguez-R L M, Pizarro V, et al. Shifts in bacterial communities of two Caribbean reef-building coral species affected by white plague disease[J]. The ISME Journal, 2012, 6(3): 502−512. doi: 10.1038/ismej.2011.123 [46] Yellowlees D, Rees T A V, Leggat W. Metabolic interactions between algal symbionts and invertebrate hosts[J]. Plant, Cell & Environment, 2008, 31(5): 679-694. [47] Hördt A, López M G, Meier-Kolthoff J P, et al. Analysis of 1, 000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria[J]. Frontiers in Microbiology, 2020, 11: 468. doi: 10.3389/fmicb.2020.00468 [48] McDevitt-Irwin J M, Baum J K, Garren M, et al. Responses of coral-associated bacterial communities to local and global stressors[J]. Frontiers in Marine Science, 2017, 4: 262. doi: 10.3389/fmars.2017.00262 [49] O'Brien P A, Morrow K M, Willis B L, et al. Implications of ocean acidification for marine microorganisms from the free-living to the host-associated[J]. Frontiers in Marine Science, 2016, 3: 47. [50] Hayashi K, Senuma W, Kai K, et al. Major exopolysaccharide, EPS I, is associated with the feedback loop in the quorum sensing of Ralstonia solanacearum strain OE1-1[J]. Molecular Plant Pathology, 2019, 20(12): 1740−1747. doi: 10.1111/mpp.12870 [51] Ainsworth T D, Krause L, Bridge T, et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts[J]. The ISME Journal, 2015, 9(10): 2261−2274. doi: 10.1038/ismej.2015.39 [52] Zhou Jin, Lin Zijun, Cai Zhonghua, et al. Opportunistic bacteria use quorum sensing to disturb coral symbiotic communities and mediate the occurrence of coral bleaching[J]. Environmental Microbiology, 2020, 22(5): 1944−1962. doi: 10.1111/1462-2920.15009 [53] Teplitski M, Warriner K, Bartz J, et al. Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety[J]. Trends in Microbiology, 2011, 19(3): 121−127. doi: 10.1016/j.tim.2010.11.007 -



 下载:
下载: