南极红藻Iridaea cordata和Curdiea racovitzae转录组分析及其极端光环境适应相关基因的挖掘
doi: 10.3969/j.issn.0253-4193.2020.10.011
De nova transcriptome analysis and mining extreme light environments acclimation responding genes of Antarctic seaweed Iridaea cordata (Gigartinales, Rhodophyta) and Curdiea racovitzae (Gracilariaceae, Rhodophyta)
-
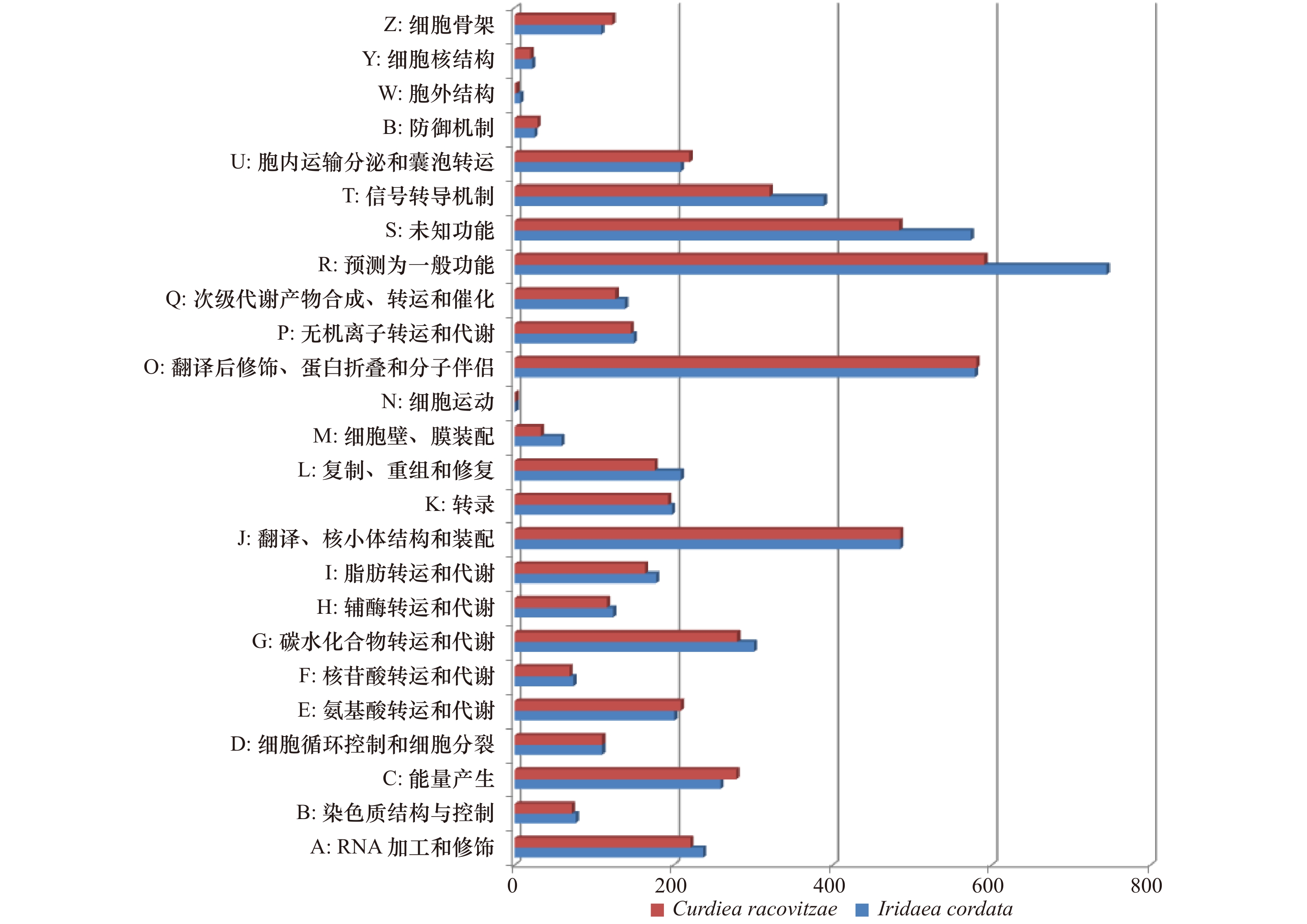
摘要: 南极红藻具有重要的生态学功能和开发利用价值。南极极端环境赋予了其独特的生理耐受机制,也是发现新基因和代谢途径的理想材料。我们测序分析了南极产胶红藻Iridaea cordata (Turner) Bory和Curdiea racovitzae Hariot的转录组序列,并与其常温近缘种进行了比较,同时挖掘了其与光限制和强紫外线辐射等光环境适应相关的基因。I. cordata和C. racovitzae的转录组序列分别拼接成了14055条和12006条非冗余基因,平均长度分别为1473 bp和1448 bp。在I. cordata转录组中发现多条与绿藻基因同源的捕光复合物LHC基因Lhca2、Lhca6和Lhcb,并且在两种南极红藻中都各发现了1条编码结合岩藻黄质和Chl a/c蛋白的Lhcf基因,目前尚未在其他红藻中发现这类基因。光裂解酶修复紫外线诱导DNA损伤,在I. cordata的转录组序列中发现了6−4光裂解酶,光裂解酶CPD I和CPD II基因,而C. racovitzae转录组序列中仅找到了光裂解酶CPD II基因。尽管南极红藻中这些特有基因的功能有待进一步的验证,但是本文为后续研究红藻的南极极端光环境适应机制提供了方向。
-
关键词:
- 南极红藻 /
- 转录组 /
- 极端光环境适应 /
- Iridaea cordata /
- Curdiea racovitzae
Abstract: Antarctic red algae play important roles in the coastal ecosystems and industrial applications. Meanwhile, their unique physiological acclimation mechanisms to the extreme environments endow them to be ideal organisms for discovering new genes and new metabolic pathways. In this study, we sequenced the transcriptomes of Antarctic red algae Iridaea cordata (Turner) Bory and Curdiea racovitzae Hariot, and compared with their moderate temperature close relatives. The transcriptome sequences of I. cordata and C. racovitzae were assembled into 14055 and 12006 Unigenes, with an average length of 1473 bp and 1448 bp, respectively. The Lhca2, Lhca6 and Lhcb genes homologous to the green algae genes were found in I. cordata transcriptome while not in other red algae. Lhcf gene encoding fucoxanthine and Chl a/c binding protein presenting in brown algae and diatoms were identified in both I. cordata and C. racovitzae. Photolyase repairs UV-induced DNA damages. 6-4 photolyase, CPD I and CPD II genes were identified in the transcriptome of I. cordata, while only CPD II gene was found in the transcriptome of C. racovitzae. Although the functions of those specific genes in Antarctic red algae are expected further investigation, our study provides a foundation for the following researches on the acclimation mechanisms of seaweeds to the extreme light environments in Antarctica. -
图 3 角叉菜Chondrus crispus与Iridaea cordata,龙须菜Gracilariopsis chorda与Curdiea racovitzae匹配基因数目的韦恩图
红色表示南极红藻,绿色表示常温红藻
Fig. 3 Diagram of the matching gene numbers between Chondrus crispus and Iridaea cordata, Gracilariopsis chorda and Curdiea racovitzae
Red represents the Antarctic red algae, green represents the normal temperate red algae
图 4 捕光复合物的系统进化树
进化树由来自几种主要红藻的基因组或转录组中的所有LHC基因编码的蛋白序列,以及褐藻、绿藻和硅藻的部分LHC蛋白序列组成。5种类型的LHC用不同颜色的方框区分。LHCR为红藻型捕光复合物,LHCF是硅藻和褐藻中主要的天线蛋白,LHCa2、LHCa6和LHCb主要来自绿藻。IcoTRINITY和CraTRINITY分别表示Iridaea cordata和Curdiea racovitzae的蛋白编码序列。进化树经MEGA-X软件用邻接法构建,得到1 000次检验一致性的系统树,自举值在相应的节点标示
Fig. 4 Phylogenetic tree of the light harvesting complex superfamily
All of LHC sequences from the genome or transcriptome of several red algae species, and some LHCs from brown algae, green algae and diatoms are shown in the tree. Five LHC branches are represented by different color boxes. LHCR is from red algae, LHCf is mainly from diatom and brown algae, LHCa2, LHCa6 and LHCb are mainly from green algae. IcoTRINITY and CraTRINITY represent the sequences from Iridaea cordata and Curdiea racovitzae, respectively. The phylogenetic tree is obtained using Neighbor-Joining (NJ) method by MEGA-X software. Bootstrap values referred to 1 000 replicates are shown at the nodes
图 5 Iridaea cordata的Lhca2、Lhca6和Lhcb基因编码的蛋白序列与绿藻LHC序列的比较
3种LHC序列用绿色线段区分,*表示参考莱茵衣藻序列中的叶绿素分子结合位点。Ath:拟南芥;Cin:衣藻;Cre:莱茵衣藻;Csu:胶球藻;Cva:小球藻;Dsa:杜氏盐藻;Ico:南极红藻;Ota:绿藻;Vca:团藻
Fig. 5 Alignment of Lhca2, Lhca6 and Lhcb of Iridaea cordata with homologous LHCs in green algae
Three LHC groups are distinguished with green lines; *marks amino acid residues that binding chlorophylls in Chlamydomonas reinhardtii sequences. Ath: Arabidopsis thaliana; Cin: Chlamydomonas incerta; Cre: Chlamydomonas reinhardtii; Csu: Coccomyxa subellipsoidea; Cva: Chlorella variabilis; Dsa: Dunaliella salina; Ico: Iridaea cordata; Ota: Ostreococcus tauri; Vca: Volvox carteri f. nagariensis
图 6 光裂解酶/隐花色素超家族的系统进化树
进化树中序列的物种包括红藻(红色标记),绿藻和高等植物(绿色标记)以及褐藻和硅藻(褐色标记)。两株南极红藻用红线框标记。聚类的6个分支用不同颜色的边线表示,包括环丁烷嘧啶二聚体光裂解酶CPDI和CPDII,6−4嘧啶酮光裂解酶,隐花色素Cry,Cry-DASH和Cry-DASH II。1 000次检验一致性的系统树经MEGA-X软件用邻接法构建,自举值在相应的节点标示。Ath: 拟南芥; Ccr: 皱波角叉菜; Cme: 红藻; Cra: 南极红藻; Cre: 莱茵衣藻; Esi: 长囊水云; Gch: 绳状龙须菜; Ico: 南极红藻; Gsu: 红藻; Ptr: 三角褐指藻; Tps: 伪矮海链藻
Fig. 6 Phylogenetic tree of the cryptochrome/photolyase family
Species in the evolutionary tree include red algae (red marker), green algae and higher plants (green marker), and brown algae and diatoms (brown marker). Two Antarctic red algae are labeled with red frame. The six clusters are represented by different color lines, including photolyase CPD I and CPD II, 6−4 photolyase, and cryptochromes Cry, Cry-DASH and Cry-DASH II. The phylogenetic tree is constructed using Neighbor-Joining (NJ) method by MEGA-X. Bootstrap values referred to 1000 replicates are shown at the nodes. Taxonomic abbreviations: Ath: Arabidopsis thaliana; Ccr: Chondrus crispus; Cme: Cyanidioschyzon merolae; Cra: Curdiea racovitzae; Cre: Chlamydomonas reinhardtii; Esi: Ectocarpus siliculosus; Gch: Gracilariopsis chorda; Ico: Iridaea cordata; Gsu: Galdieria sulphuraria; Ptr: Phaeodactylum tricornutum; Tps: Thalassiosira pseudonana
表 1 南极红藻Iridaea cordata和Curdiea racovitzae转录本和非冗余基因
Tab. 1 The contigs and unigenes of Iridaea cordata and Curdiea racovitzae
Iridaea cordata Curdiea racovitzae 重叠群 非冗余基因 重叠群 非冗余基因 总长度/bp 23957720 20707433 19982213 17378947 序列数目 32395 14055 26226 12006 最大序列长度/bp 24631 24418 27865 26227 平均序列长度/bp 740 1473 762 1448 N50/bp 2724 3133 3256 3673 N50 序列数 2329 1901 1635 1307 N90/bp 199 537 210 457 N90 序列数 15816 7637 12822 6213 GC% 53.82 53.29 47.28 46.88 表 2 Iridaea cordata和Curdiea racovitzae转录组序列在各个数据库的注释结果
Tab. 2 The annotation results of Iridaea cordata and Curdiea racovitzae transcriptome
数据库 Iridaea cordata Curdiea racovitzae 数目 百分比 数目 百分比 NR 4788 34.07 4468 37.21 GO 1806 12.85 1838 15.31 KEGG 2024 14.40 1903 15.85 EggNOG 4523 32.18 4238 35.3 Swissprot 4864 34.61 4938 41.13 全部数据库 972 6.92 947 7.89 -
[1] Wiencke C, Clayton M. Antarctic seaweeds[M]//Wägele J W. Synopsis of the Antarctic Benthos. Ruggell: ARG Gantner, 2002. [2] Oliveira E C, Absher T M, Pellizzari F M, et al. The seaweed flora of Admiralty Bay, King George Island, Antarctic[J]. Polar Biology, 2009, 32(11): 1639−1647. doi: 10.1007/s00300-009-0663-9 [3] Pellizzari F, Silva M C, Silva E M, et al. Diversity and spatial distribution of seaweeds in the South Shetland Islands, Antarctica: an updated database for environmental monitoring under climate change scenarios[J]. Polar Biology, 2017, 40(8): 1671−1685. doi: 10.1007/s00300-017-2092-5 [4] Martín A, Miloslavich P, Díaz Y, et al. Intertidal benthic communities associated with the macroalgae Iridaea cordata and Adenocystis utricularis in King George Island, Antarctica[J]. Polar Biology, 2016, 39(2): 207−220. doi: 10.1007/s00300-015-1773-1 [5] Dhargalkar V K, Verlecar X N. Southern Ocean seaweeds: a resource for exploration in food and drugs[J]. Aquaculture, 2009, 287(3/4): 229−242. [6] Guillemin M L, Dubrasquet H, Reyes J, et al. Comparative phylogeography of six red algae along the Antarctic Peninsula: extreme genetic depletion linked to historical bottlenecks and recent expansion[J]. Polar Biology, 2018, 41(5): 827−837. doi: 10.1007/s00300-017-2244-7 [7] McCandless E L, Craigie J S, Hansen J E. Carrageenans of gametangial and tetrasporangial stages of Iridaea cordata (Gigartinaceae)[J]. Canadian Journal of Botany, 1975, 53(20): 2315−2318. doi: 10.1139/b75-256 [8] Kim H J, Kim W J, Koo B W, et al. Anticancer activity of sulfated polysaccharides isolated from the Antarctic red seaweed Iridaea cordata[J]. Ocean and Polar Research, 2016, 38(2): 129−137. doi: 10.4217/OPR.2016.38.2.129 [9] Falshaw R, Furneaux R H, Stevenson D E. Agars from nine species of red seaweed in the genus Curdiea (Gracilariaceae, Rhodophyta)[J]. Carbohydrate Research, 1998, 308(1/2): 107−115. [10] Wiencke C, Rahmel J, Karsten U, et al. Photosynthesis of marine Macroalgae from Antarctica: light and temperature requirements[J]. Botanica Acta, 1993, 106(1): 78−87. doi: 10.1111/j.1438-8677.1993.tb00341.x [11] Navarro N P, Mansilla A, Plastino E M. UVB radiation induces changes in the ultra-structure of Iridaea cordata[J]. Micron, 2010, 41(7): 899−903. doi: 10.1016/j.micron.2010.06.004 [12] Navarro N P, Huovinen P, Gómez I. Stress tolerance of Antarctic macroalgae in the early life stages[J]. Revista Chilena de Historia Natural, 2016, 89(1): 5. doi: 10.1186/s40693-016-0051-0 [13] McClintock J B, Karentz D. Mycosporine-like amino acids in 38 species of subtidal marine organisms from McMurdo Sound, Antarctica[J]. Antarctic Science, 1997, 9(4): 392−398. doi: 10.1017/S0954102097000503 [14] Cormaci M, Furnari G, Scammacca B, et al. Summer biomass of a population of Iridaea cordata (Gigartinaceae, Rhodophyta) from Antarctica[J]. Hydrobiologia, 1996, 326−327(1): 267−272. doi: 10.1007/BF00047817 [15] Wiencke C, Clayton M N, Gómez I, et al. Life strategy, ecophysiology and ecology of seaweeds in polar waters[J]. Reviews in Environmental Science and Bio/Technology, 2007, 6(1/3): 95−126. [16] Wulff A, Iken K, Liliana Quartino M, et al. Biodiversity, biogeography and zonation of marine benthic micro- and macroalgae in the Arctic and Antarctic[J]. Botanica Marina, 2009, 52(6): 491−507. [17] Amsler D C, Okogbue I N, Landry D M, et al. Potential chemical defenses against diatom fouling in Antarctic macroalgae[J]. Botanica Marina, 2005, 48(4): 318−322. [18] Martins R M, Nedel F, Guimarães V B S, et al. Macroalgae extracts from Antarctica have antimicrobial and anticancer potential[J]. Frontiers in Microbiology, 2018, 9: 412. doi: 10.3389/fmicb.2018.00412 [19] Maccoll R, Eisele L E, Williams E C, et al. The discovery of a novel R-phycoerythrin from an Antarctic red alga[J]. Journal of Biological Chemistry, 1996, 271(29): 17157−17160. doi: 10.1074/jbc.271.29.17157 [20] Zacher K, Roleda M Y, Wulff A, et al. Responses of Antarctic Iridaea cordata (Rhodophyta) tetraspores exposed to ultraviolet radiation[J]. Phycological Research, 2009, 57(3): 186−193. doi: 10.1111/j.1440-1835.2009.00538.x [21] Carvalho E L, Maciel L F, Macedo P E, et al. De novo assembly and annotation of the Antarctic alga Prasiola crispa Transcriptome[J]. Frontiers in Molecular Biosciences, 2017, 4: 89. [22] Grabherr M G, Haas B J, Yassour M, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome[J]. Nature Biotechnology, 2011, 29(7): 644−652. doi: 10.1038/nbt.1883 [23] Powell S, Szklarczyk D, Trachana K, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges[J]. Nucleic Acids Research, 2012, 40(D1): D284−D289. doi: 10.1093/nar/gkr1060 [24] Moriya Y, Itoh M, Okuda S, et al. KAAS: an automatic genome annotation and pathway reconstruction server[J]. Nucleic Acids Research, 2007, 35(S2): W182−W185. [25] Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547−1549. doi: 10.1093/molbev/msy096 [26] Collén J, Porcel B, Carré W, et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(13): 5247−5252. doi: 10.1073/pnas.1221259110 [27] Sun X, Wu J, Wang G C, et al. Genomic analyses of unique carbohydrate and phytohormone metabolism in the macroalga Gracilariopsis lemaneiformis (Rhodophyta)[J]. BMC Plant Biology, 2018, 18(1): 94. doi: 10.1186/s12870-018-1309-2 [28] Choi S, Hwang M S, Im S, et al. Transcriptome sequencing and comparative analysis of the gametophyte thalli of Pyropia tenera under normal and high temperature conditions[J]. Journal of Applied Phycology, 2013, 25(4): 1237−1246. doi: 10.1007/s10811-012-9921-2 [29] Im S, Choi S, Hwang M S, et al. De novo assembly of transcriptome from the gametophyte of the marine red algae Pyropia seriata and identification of abiotic stress response genes[J]. Journal of Applied Phycology, 2015, 27(3): 1343−1353. doi: 10.1007/s10811-014-0406-3 [30] Green B R, Durnford D G. The chlorophyll-carotenoid proteins of oxygenic photosynthesis[J]. Annual Review of Plant Physiology and Plant Molecular Biology, 1996, 47: 685−714. doi: 10.1146/annurev.arplant.47.1.685 [31] Engelken J, Brinkmann H, Adamska I. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily[J]. BMC Evolutionary Biology, 2010, 10(1): 233. doi: 10.1186/1471-2148-10-233 [32] Durnford D G, Deane J A, Tan S, et al. A phylogenetic assessment of the eukaryotic light-harvesting antenna proteins, with implications for plastid evolution[J]. Journal of Molecular Evolution, 1999, 48(1): 59−68. doi: 10.1007/PL00006445 [33] Busch A, Hippler M. The structure and function of eukaryotic photosystem I[J]. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2011, 1807(8): 864−877. doi: 10.1016/j.bbabio.2010.09.009 [34] de Martino A, Douady D, Quinet-Szely M, et al. The light-harvesting antenna of brown algae: highly homologous proteins encoded by a multigene family[J]. European Journal of Biochemistry, 2000, 267(17): 5540−5549. doi: 10.1046/j.1432-1327.2000.01616.x [35] Reith M. Molecular biology of Rhodophyte and Chromophyte plastids[J]. Annual Review of Plant Physiology and Plant Molecular Biology, 2014, 46: 549−575. [36] Ballottari M, Girardon J, Dall’Osto L, et al. Evolution and functional properties of Photosystem II light harvesting complexes in eukaryotes[J]. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2012, 1817(1): 143−157. doi: 10.1016/j.bbabio.2011.06.005 [37] Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors[J]. Chemical Reviews, 2003, 103(6): 2203−2237. doi: 10.1021/cr0204348 [38] Chaves I, Pokorny R, Byrdin M, et al. The cryptochromes: blue light photoreceptors in plants and animals[J]. Annual Review of Plant Biology, 2011, 62: 335−364. doi: 10.1146/annurev-arplant-042110-103759 [39] Selby C P, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(47): 17696−17700. doi: 10.1073/pnas.0607993103 [40] Fortunato A E, Annunziata R, Jaubert M, et al. Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms[J]. Journal of Plant Physiology, 2015, 172: 42−54. doi: 10.1016/j.jplph.2014.06.011 [41] Varshney R K, Graner A, Sorrells M E. Genic microsatellite markers in plants: features and applications[J]. Trends in Biotechnology, 2005, 23(1): 48−55. doi: 10.1016/j.tibtech.2004.11.005 [42] Allcock A L, Strugnell J M. Southern Ocean diversity: new paradigms from molecular ecology[J]. Trends in Ecology & Evolution, 2012, 27(9): 520−528. [43] Riesgo A, Taboada S, Avila C. Evolutionary patterns in Antarctic marine invertebrates: an update on molecular studies[J]. Marine Genomics, 2015, 23: 1−13. doi: 10.1016/j.margen.2015.07.005 -




 下载:
下载: