Cloning of the McNF-κB gene of Mytilus coruscus and its role in development
-
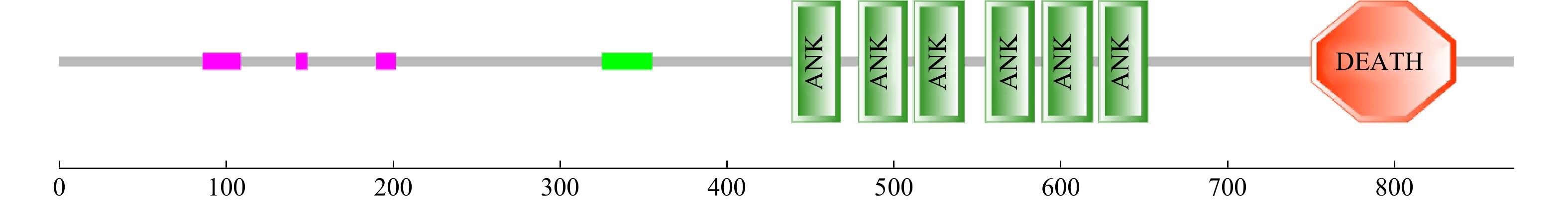
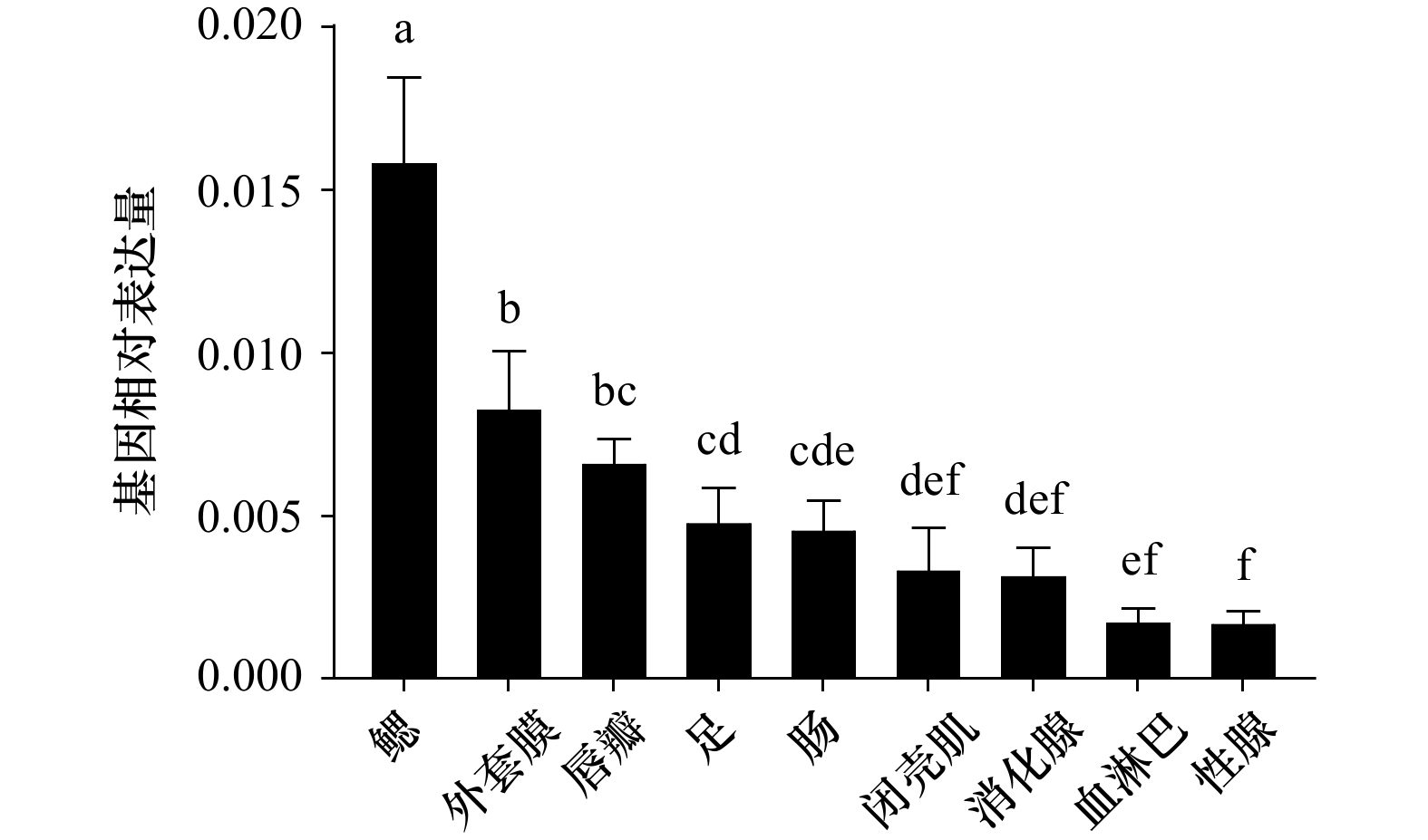
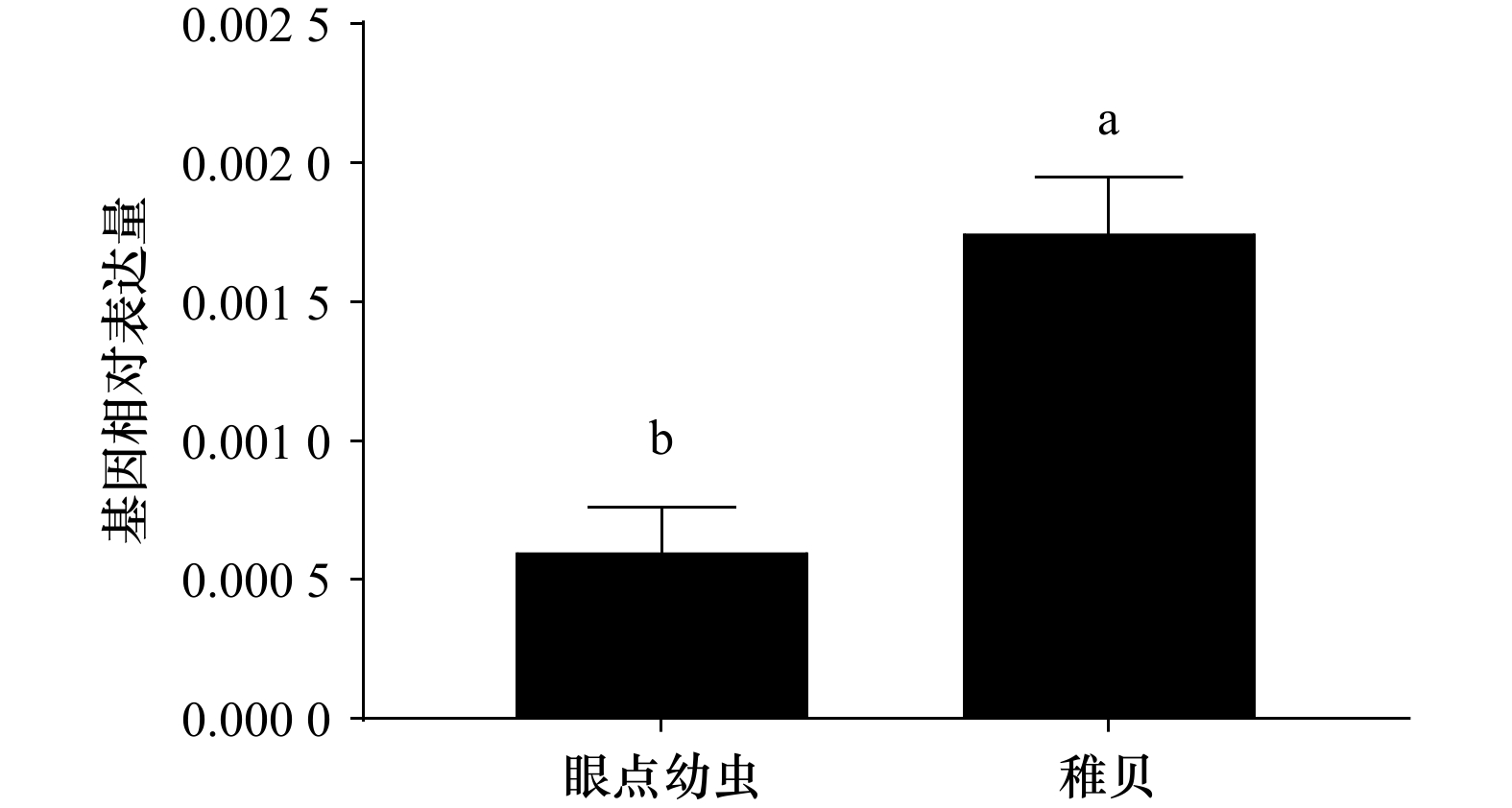
摘要: 核因子κB(Nuclear Factor kappa-B,NF-κB)具有免疫、炎症、凋亡、细胞增殖和发育的调节作用,目前NF-κB在脊椎动物和果蝇中的研究较为丰富,在贝类中的报道较少。为进一步探究NF-κB在厚壳贻贝(Mytilus coruscus)免疫和发育中的作用,本研究克隆了厚壳贻贝McNF-κB基因的序列全长,其全长为4 087 bp,开放阅读框为2 613 bp,编码871个氨基酸,具有典型的锚蛋白重复序列(ankyrinrepeat, ANK)结构域和死亡结构域。氨基酸序列分析结果发现,该基因与欧洲贻贝(Mytilus edulis)和地中海贻贝(Mytilus galloprovincialis)分别具有72.76%和66.58%同源性,且在系统进化树中与欧洲贻贝和地中海贻贝聚为一支。经实时荧光定量PCR(qRT-PCR)技术检验表明,McNF-κB基因于厚壳贻贝各组织均有分布,在鳃中表达最高;McNF-κB基因在厚壳贻贝眼点幼虫阶段和稚贝阶段均有表达,且在稚贝阶段表达量显著高于眼点幼虫阶段。利用RNA干扰技术沉默眼点幼虫McNF-κB基因后幼虫变态率显著下降,推测McNF-κB基因调控厚壳贻贝幼虫变态过程。本研究为探究McNF-κB基因如何调控厚壳贻贝发育奠定了基础。Abstract: Nuclear factor κB (NF-κB) can regulate immunity, inflammation, apoptosis, cell proliferation, and organism development. At present, NF-κB has been well studied in vertebrates and fruit flies, while its role in shellfish is still elusive. In order to further explore the role of NF-κB in the immunity and development of mussel Mytilus coruscus, the full length McNF-κB cDNA sequence was cloned from M. coruscus. McNF-κB gene was 4087 bp long, and the open reading frame was 2 613 bp, encoding 871 amino acids and had a typical ankyrinrepeat (ANK) domain and DEATH domain. The results of amino acid sequence analysis showed that the gene had 72.76% homology with M. edulis and 66.58% with M. galloprovincialis, respectively, and was clustered with M. edulis and M. galloprovincialis in the phylogenetic tree. The real-time PCR (qRT-PCR) technology showed that the McNF-κB gene was expressed distributed in all tissues of M. coruscus, and the expression was the highest in the gill. McNF-κB gene was expressed in both the pediveliger larvae stage and the juvenile stage of M. coruscus, and the expression was significantly higher in the pediveliger larvae stage than that in the juvenile stage. After using RNA interference technology to silence the McNF-κB gene of pediveliger larvae, the larval metamorphosis rate decreased significantly, indicating that this gene was involved in regulating the metamorphosis process of M. coruscus. This study provides a basis for exploring how McNF-κB gene regulates development of M. coruscus.
-
Key words:
- Mytilus coruscus /
- NF-κB /
- gene cloning /
- larval metamorphosis
-
图 1 McNF-κB基因cDNA全长及推导的氨基酸序列
下划线表示起始密码子ATG;“*”表示终止密码子;灰色区域表示ANK结构域;黑色方框表示死亡结构域
Fig. 1 Full-length cDNA and deduced amino acid sequences of McNF-κB gene
The start codon ATG is underlined; the stop codon is indicated by an asterisk;the gray area represents the ANK domain; the black box represents the DEATH domain
图 3 不同物种NF-κB氨基酸序列多重比对分析
绿色框线表示具有多个ANK结构域;红色框线表示死亡结构域;深色背景中的氨基酸序列具有100%的同源性;粉色背景中氨基酸具有大于75%的同源性;绿色背景中氨基酸具有大于50%的同源性
Fig. 3 Multiple alignment analysis of NF-κB amino acid sequences of different species
Green borders indicate multiple ANK domains; red borders indicate DEATH domain; the amino acids in dark gray boxes indicate conserved residues with 100% homology; the amino acids in pink boxes show conservation of residues with > 75% homology; the amino acids in green boxes show conservation of residues with > 50% homology
表 1 厚壳贻贝NF-κB基因cDNA全长克隆和mRNA表达分析所用的引物序列
Tab. 1 Primers sequences used for full-length cDNA cloning and Mytilus coruscus NF-κB gene mRNA expression analysis
引物名称 序列 用途 McNF-κB-1-3′ RACE TGACAGAAAAGGCAATACCCCG 3′ RACE McNF-κB-2-3′ RACE GAGGTGACCCTGAGATGGAA 3′ RACE McNF-κB-1-5′ RACE GAGGTTGGCAAAAGTGACAG 5′ RACE McNF-κB-2-5′ RACE CTTCAGAACATTTGCCCCC 5′ RACE McNF-κB-RT-F GTATACCCAGACCCCAATC qRT-PCR McNF-κB-RT-R TCTTCTACCGTCACCACC qRT-PCR EF-1α-RT-F CACCACGAGTCTCTCCCTGA qRT-PCR EF-1α-RT-R GCTGTCACCACAGACCA TTCC qRT-PCR -
[1] 杨金龙, 李树恒, 刘志伟, 等. 厚壳贻贝胚胎和早期幼虫神经系统发育的初步研究[J]. 水产学报, 2013, 37(4): 512−519. doi: 10.3724/SP.J.1231.2013.38433Yang Jinlong, Li Shuheng, Liu Zhiwei, et al. Primary study on neuronal development of the embryo and early larvae of the mussel Mytilus coruscus[J]. Journal of Fisheries of China, 2013, 37(4): 512−519. doi: 10.3724/SP.J.1231.2013.38433 [2] Yang Jinlong, Li Shuheng, Li Yifeng, et al. Effects of neuroactive compounds, ions and organic solvents on larval metamorphosis of the mussel Mytilus coruscus[J]. Aquaculture, 2013, 396−399: 106−112. doi: 10.1016/j.aquaculture.2013.02.039 [3] 王朝新. 厚壳贻贝苗种规模化繁育技术[J]. 现代农业科技, 2021(17): 208−210.Wang Chaoxin. Large-scale breeding technology of the mussel Mytilus coruscus seeds[J]. Modern Agricultural Science and Technology, 2021(17): 208−210. [4] Wang Chong, Bao Weiyang, Gu Zhongqi, et al. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to natural biofilms[J]. Biofouling, 2012, 28(3): 249−256. doi: 10.1080/08927014.2012.671303 [5] 刘钰珠. Toll样受体在厚壳贻贝免疫和发育中的作用研究[D]. 上海: 上海海洋大学, 2019.Liu Yuzhu. Effect of Toll-like receptor on immunity and development of Mytilus coruscus[D]. Shanghai: Shanghai Ocean University, 2019. [6] 徐跃峰, 李一峰, 梁箫, 等. 厚壳贻贝Wnt4基因时空表达[J]. 水产学报, 2016, 40(10): 1567−1575.Xu Yuefeng, Li Yifeng, Liang Xiao, et al. Temporal-spatial expression of Wnt4 gene in the mussel Mytilus coruscus[J]. Journal of Fisheries of China, 2016, 40(10): 1567−1575. [7] Xu Yuefeng, Liang Xiao, Chen Yuru, et al. Wnt7b gene expression and functional analysis in the mussel Mytilus coruscus[J]. Genetics and Molecular Research, 2016, 15(4): gmr15048866. [8] Zhu Youting, Zhang Ya, Liu Yuzhu, et al. Nitric oxide negatively regulates larval metamorphosis in hard-shelled mussel (Mytilus coruscus)[J]. Frontiers in Marine Science, 2020, 7: 356. doi: 10.3389/fmars.2020.00356 [9] Yuk J M, Jo E K. Toll-like receptors and innate immunity[J]. Journal of Bacteriology and Virology, 2011, 41(4): 225−235. doi: 10.4167/jbv.2011.41.4.225 [10] Du Qiang, Geller D. Cross-regulation between Wnt and NF-κB signaling pathways[J]. Forum on Immunopathological Diseases and Therapeutics, 2010, 1(3): 155−181. doi: 10.1615/ForumImmunDisTher.v1.i3.10 [11] Zinatizadeh M R, Schock B, Chalbatani G M, et al. The Nuclear Factor Kappa B (NF-κB) signaling in cancer development and immune diseases[J]. Genes & Diseases, 2021, 8(3): 287−297. [12] Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences[J]. Cell, 1986, 46(5): 705−716. doi: 10.1016/0092-8674(86)90346-6 [13] Ghosh S, May M J, Kopp E B. NF-κB and REL proteins: evolutionarily conserved mediators of immune responses[J]. Annual Review of Immunology, 1998, 16: 225−260. doi: 10.1146/annurev.immunol.16.1.225 [14] Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism[J]. Cell, 1986, 47(6): 921−928. doi: 10.1016/0092-8674(86)90807-X [15] 孙伟东, 张志强, 朴大勋. NF-κB信号通路抑制结肠炎及炎症相关结肠癌的研究[J]. 医学综述, 2020, 26(8): 1521−1525.Sun Weidong, Zhang Zhiqiang, Piao Daxun. Research progress of NF-κB signaling pathway inhibition on colitis and inflammation-associated colon cancer[J]. Medical Recapitulate, 2020, 26(8): 1521−1525. [16] Govind S. Control of development and immunity by Rel transcription factors in Drosophila[J]. Oncogene, 1999, 18(49): 6875−6887. doi: 10.1038/sj.onc.1203223 [17] Kleino A, Ramia N F, Bozkurt G, et al. Peptidoglycan-sensing receptors trigger the formation of functional amyloids of the adaptor protein Imd to initiate Drosophila NF-κB signaling[J]. Immunity, 2017, 47(4): 635−647.e6. doi: 10.1016/j.immuni.2017.09.011 [18] Rusch J, Levine M. Regulation of the dorsal morphogen by the Toll and torso signaling pathways: a receptor tyrosine kinase selectively masks transcriptional repression[J]. Genes & Development, 1994, 8(11): 1247−1257. [19] Barillas-Mury C, Charlesworth A, Gross I, et al. Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae[J]. The EMBO Journal, 1996, 15(17): 4691−4701. doi: 10.1002/j.1460-2075.1996.tb00846.x [20] Yu Aiqing, Jin Xingkun, Li Shuang, et al. Molecular cloning and expression analysis of a dorsal homologue from Eriocheir sinensis[J]. Developmental & Comparative Immunology, 2013, 41(4): 723−727. [21] Shin S W, Kokoza V, Lobkov I, et al. Relish-mediated immune deficiency in the transgenic mosquito Aedes aegypti[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(5): 2616−2621. [22] Shin S W, Kokoza V, Bian Guowu, et al. REL1, a homologue of Drosophila dorsal, regulates Toll antifungal immune pathway in the female mosquito Aedes aegypti[J]. Journal of Biological Chemistry, 2005, 280(16): 16499−16507. doi: 10.1074/jbc.M500711200 [23] Huang Xiande, Yin Zhixin, Liao Jixiang, et al. Identification and functional study of a shrimp Relish homologue[J]. Fish & Shellfish Immunology, 2009, 27(2): 230−238. [24] Huang Xiande, Yin Zhixin, Jia Xiaoting, et al. Identification and functional study of a shrimp Dorsal homologue[J]. Developmental & Comparative Immunology, 2010, 34(2): 107−113. [25] 梁芳梅, 朱鹏, 邱春桃, 等. 长毛明对虾核转录因子NF-κB家族基因的克隆及在细菌侵染过程中的表达变化[J]. 中国水产科学, 2022, 29(11): 1551−1563.Liang Fangmei, Zhu Peng, Qiu Chuntao, et al. Cloning and expression analysis of NF-κB family genes under bacterial infection of Fenneropenaeus penicillatus[J]. Journal of Fishery Sciences of China, 2022, 29(11): 1551−1563. [26] 赵姣姣, 陶震, 周素明, 等. 三疣梭子蟹NF-κB家族基因Relish和Dorsal的克隆及表达特征[J]. 水生生物学报, 2019, 43(2): 298−304.Zhao Jiaojiao, Tao Zhen, Zhou Suming, et al. Cloning and expression of NF-κB family genes in portunus trituberculatus[J]. Acta Hydrobiologica Sinica, 2019, 43(2): 298−304. [27] Huang Xiande, Liu Wenguang, Guan Yunyan, et al. Molecular cloning and characterization of class I NF-κB transcription factor from pearl oyster (Pinctada fucata)[J]. Fish & Shellfish Immunology, 2012, 33(3): 659−666. [28] Wu Xi, Xiong Xunhao, Xie Liping, et al. Pf-rel, a rel/nuclear factor-κB homolog identified from the pearl oyster, Pinctada fucata[J]. Acta Biochimica et Biophysica Sinica, 2007, 39(7): 533−539. doi: 10.1111/j.1745-7270.2007.00306.x [29] Montagnani C, Kappler C, Reichhart J M, et al. Cg-Rel, the first Rel/NF-κB homolog characterized in a mollusk, the Pacific oyster Crassostrea gigas[J]. FEBS Letters, 2004, 561(1/3): 75−82. [30] Jiang Yusheng, Wu Xinzhong. Characterization of a Rel\NF-κB homologue in a gastropod abalone, Haliotis diversicolor supertexta[J]. Developmental & Comparative Immunology, 2007, 31(2): 121−131. [31] De Zoysa M, Nikapitiya C, Oh C, et al. Molecular evidence for the existence of lipopolysaccharide-induced TNF-α factor (LITAF) and Rel/NF-kB pathways in disk abalone (Haliotis discus discus)[J]. Fish & Shellfish Immunology, 2010, 28(5/6): 754−763. [32] Zhang Siming, Coultas K A. Identification and characterization of five transcription factors that are associated with evolutionarily conserved immune signaling pathways in the schistosome-transmitting snail Biomphalaria glabrata[J]. Molecular Immunology, 2011, 48(15/16): 1868−1881. [33] Zhou Zhi, Wang Mengqiang, Zhao Jianmin, et al. The increased transcriptional response and translocation of a Rel/NF-κB homologue in scallop Chlamys farreri during the immune stimulation[J]. Fish & Shellfish Immunology, 2013, 34(5): 1209−1215. [34] Yang Jinlong, Li Yifeng, Bao Weiyang, et al. Larval metamorphosis of the mussel Mytilus galloprovincialis Lamarck, 1819 in response to neurotransmitter blockers and tetraethylammonium[J]. Biofouling, 2011, 27(2): 193−199. doi: 10.1080/08927014.2011.553717 [35] Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547−1549. doi: 10.1093/molbev/msy096 [36] Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences[J]. Journal of Molecular Evolution, 1980, 16(2): 111−120. doi: 10.1007/BF01731581 [37] 梁箫, 陈珂, 陈艳文, 等. 厚壳贻贝5-羟色胺2A受体(5-HT2AR)基因克隆和时空表达[J]. 水产学报, 2018, 42(12): 1869−1879.Liang Xiao, Chen Ke, Chen Yanwen, et al. 5-HT2AR gene clone and temporal-spatial expression in the mussel Mytilus coruscus[J]. Journal of Fisheries of China, 2018, 42(12): 1869−1879. [38] Li Yifeng, Cheng Yulan, Chen Ke, et al. Thyroid hormone receptor: a new player in epinephrine-induced larval metamorphosis of the hard-shelled mussel[J]. General and Comparative Endocrinology, 2020, 287: 113347. doi: 10.1016/j.ygcen.2019.113347 [39] Liang Xiao, Chen Ke, Li Yifeng, et al. An ɑ2-adrenergic receptor is involved in larval metamorphosis in the mussel, Mytilus coruscus[J]. Biofouling, 2019, 35(9): 986−996. doi: 10.1080/08927014.2019.1685661 [40] Gilmore T D. The Rel/NF-κB signal transduction pathway: introduction[J]. Oncogene, 1999, 18(49): 6842−6844. doi: 10.1038/sj.onc.1203237 [41] Manfruelli P, Reichhart J M, Steward R, et al. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF[J]. The EMBO Journal, 1999, 18(12): 3380−3391. doi: 10.1093/emboj/18.12.3380 [42] Rutschmann S, Jung A C, Hetru C, et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila[J]. Immunity, 2000, 12(5): 569−580. doi: 10.1016/S1074-7613(00)80208-3 [43] Aggarwal K, Silverman N. Positive and negative regulation of the Drosophila immune response[J]. BMB Reports, 2008, 41(4): 267−277. doi: 10.5483/BMBRep.2008.41.4.267 [44] Silverman N, Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity[J]. Genes & Development, 2001, 15(18): 2321−2342. [45] Hedengren M, BengtÅsling, Dushay M S, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila[J]. Molecular Cell, 1999, 4(5): 827−837. doi: 10.1016/S1097-2765(00)80392-5 [46] Davidson B, Swalla B J. A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response[J]. Development, 2002, 129(20): 4739−4751. doi: 10.1242/dev.129.20.4739 [47] 尹诚, 张丽莉, 王国栋, 等. 笠贝幼虫变态相关基因筛选及分子网络构建[J]. 集美大学学报(自然科学版), 2015, 20(5): 339−347.Yin Cheng, Zhang Lili, Wang Guodong, et al. Indentification of metamorphosis related genes and development of molecular networks of Lottia gigantea larval[J]. Journal of Jimei University (Natural Science), 2015, 20(5): 339−347. [48] Williams E A, Degnan B M, Gunter H, et al. Widespread transcriptional changes pre-empt the critical pelagic-benthic transition in the vetigastropod Haliotis asinina[J]. Molecular Ecology, 2009, 18(5): 1006−1025. doi: 10.1111/j.1365-294X.2008.04078.x [49] Azumi K, Sabau S V, Fujie M, et al. Gene expression profile during the life cycle of the urochordate Ciona intestinalis[J]. Developmental Biology, 2007, 308(2): 572−582. doi: 10.1016/j.ydbio.2007.05.022 [50] 刘志显, 李嘉政, 梁邻利, 等. 厚壳贻贝McCaspase 3−4基因的克隆及其在幼虫变态中的作用[J]. 水生生物学报, 2022, 46(8): 1168−1176.Liu Zhixian, Li Jiazheng, Liang Linli, et al. Molecular cloning of McCaspase 3−4 and its functions in Mytilus coruscus larval metamorphosis[J]. Acta Hydrobiologica Sinica, 2022, 46(8): 1168−1176. [51] Hayden M S, Ghosh S. Signaling to NF-κB[J]. Genes & Development, 2004, 18(18): 2195−2224. [52] Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity[J]. Trends in Immunology, 2004, 25(6): 280−288. doi: 10.1016/j.it.2004.03.008 [53] 吴静娴, 李嘉政, 胡晓梦, 等. 脂多糖对细菌生物被膜形成及厚壳贻贝幼虫变态的影响[J]. 水产学报, 2022, 46(11): 2134−2142.Wu Jingxian, Li Jiazheng, Hu Xiaomeng, et al. Effects of lipopolysaccharide on biofilm formation and larval metamorphosis of the mussel Mytilus coruscus[J]. Journal of Fisheries of China, 2022, 46(11): 2134−2142. [54] Liang Xiao, Zhang Xiukun, Peng Lihua, et al. The flagellar gene regulates biofilm formation and mussel larval settlement and metamorphosis[J]. International Journal of Molecular Sciences, 2020, 21(3): 710. doi: 10.3390/ijms21030710 [55] Chu Zhiliang, McKinsey T A, Liu L, et al. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-κB control[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(19): 10057−10062. [56] Wang Cunyu, Mayo M W, Korneluk R G, et al. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation[J]. Science, 1998, 281(5383): 1680−1683. doi: 10.1126/science.281.5383.1680 [57] Papa S, Zazzeroni F, Pham C G, et al. Linking JNK signaling to NF-κB: a key to survival[J]. Journal of Cell Science, 2004, 117(22): 5197−5208. doi: 10.1242/jcs.01483 [58] Campbell K J, Rocha S, Perkins N D. Active repression of antiapoptotic gene expression by RelA(p65) NF-κB[J]. Molecular Cell, 2004, 13(6): 853−865. doi: 10.1016/S1097-2765(04)00131-5 [59] Hettmann T, DiDonato J, Karin M, et al. An essential role for nuclear factor κB in promoting double positive thymocyte apoptosis[J]. Journal of Experimental Medicine, 1999, 189(1): 145−158. doi: 10.1084/jem.189.1.145 [60] Chen Xufeng, Kandasamy K, Srivastava R K. Differential roles of RelA (p65) and c-Rel subunits of nuclear factor κB in tumor necrosis factor-related apoptosis-inducing ligand signaling[J]. Cancer Research, 2003, 63(5): 1059−1066. [61] Kaltschmidt C, Greiner J F W, Kaltschmidt B. The transcription factor NF-κB in stem cells and development[J]. Cells, 2021, 10(8): 2042. doi: 10.3390/cells10082042 [62] Lake B B, Ford R, Kao K R. Xrel3 is required for head development in Xenopus laevis[J]. Development, 2001, 128(2): 263−273. doi: 10.1242/dev.128.2.263 -




 下载:
下载: